Every Jack has His Jill: Finding a Target for Your Combinatorial Library
Inna Slynko, Jan KF Dreher and Andreas H Göller
DOI10.21767/2470-6973.100026
Inna Slynko1,2, Jan KF Dreher1 and Andreas H Göller1*
1 Bayer Pharma AG, Medicinal Chemistry - Computational Chemistry, Wuppertal, Germany
2 Grünenthal GmbH, Computational Chemistry, Aachen, Germany
- *Corresponding Author:
- Andreas H Göller
Bayer Pharma AG, Medicinal Chemistry - Computational Chemistry, Wuppertal, Germany.
Tel: +49202365442
E-mail: andreas.goeller@bayer.com
Received Date: December 21, 2017; Accepted Date: December 26, 2017; Published Date: January 01, 2018
Citation: Slynko I, Dreher JKF, Göller AH (2018) Every Jack has His Jill: Finding a Target for Your Combinatorial Library. Chem Inform Vol. 4 No. 1:1. DOI: 10.21767/2470-6973.100026
Abstract
Pharmaceutical companies regularly run campaigns to evolve their proprietary chemical libraries which are among their most valuable assets. Ultimate goal with those library expansions is to address novel chemical space with maximal fit to pharmaceutically relevant targets which is beyond just applying property or druglikeness filters. In this work we present a structured and highly automated process to identify putative biological targets starting from any chemistry-driven virtual or existing compound library. Multiple ligand similarity searches are performed in ChEMBL ligand space, linking library compounds to targets from ChEMBL database. The results are presented to the computational chemist in a highly intuitive and interactive manner. For a set of targets selected by a scientist, holo crystal structures are automatically retrieved and prepared for docking. The cocrystallized ligand, ChEMBL compounds and combinatorial library are then docked by an automatic procedure. The scientist finally is provided with a holistic picture of library-target fit hypotheses to draw his conclusions about relevant targets, library adjustments, library re-designs and ideas for completely new virtual libraries.
Keywords
Library design; Target fishing; Automated workflow
Introduction
The chemical library belongs to the biggest research assets of any pharmaceutical company. Such screening libraries are typically between one to five million compounds [1]. Whether the full library or only subsets are tested in HTS campaigns and how such subsets are composed depends on target areas, assay designs and company’s strategy. HTS and especially in vitro and in vitro assays of individual compounds are costly in terms of substance consumption. Therefore, all libraries bleed out. Instead of resynthesizing old compounds, companies set up campaigns to evolve the libraries into new chemical space following one of three strategies, namely, buying from chemical catalogs, buying readily available proprietary compounds or designing novel proprietary chemistry. Typical design concept for novel libraries is to create structurally diverse compounds with Lipinski drug-like [2] or lead-like [3,4] properties.
Since chemical space is almost infinite with approximately 1060 compounds with a molecular weight lower than 500 Da, [5] and currently only about 10 to 20 million compounds relevant to drug discovery are covered by commercial sources and proprietary repositories, the question arises: which of numerous imaginary libraries are relevant and which not?
One way to address the question of target relevance is to start from known chemical matter and to apply core modifications like changes of ring size or type, or shifting nitrogen and functional groups. Alternatively, one can design libraries purely chemistrydriven, based on attractive chemical scaffolds, synthesis routes or concepts like escaping from flatland, [6] giving diversity and serendipity a chance. Combined with IP space analysis both routes can yield viable libraries.
We were now interested if it would be possible to find the right target or target family for a subset of our internal library designs, which were originally driven by feasible chemistry and attractive novelty. Or otherwise, if it would be possible to derive a rationale how to modify such a library design in order to tailor the respective library to a specific target or target family. We expect that a library designed with a target family in mind possesses a higher chance to hit the relevant chemical space, especially, since there are many indications for existence of privileged scaffolds [7].
We know also that computational methods, especially high throughput methods like structure- or ligand-based virtual screening or target-family likeness filters, are far from perfect and will at maximum provide certain enrichments. Therefore, we decided to combine computational methods, which provide us with a high degree of automation and throughput, with optimum use of expert knowledge and guidance. Nevertheless, we have to stress that starting libraries, as well as libraries designed with help of the process described in this article, have to be strictly novel, which requires intervention of an expert and cannot be automated.
Hence the question arises: how to find the matching target for the library, or at least for some library compounds. In the last years many researchers looked into this topic mostly from a different perspective, namely, how to control target selectivity of a lead compound and avoid adverse effects, [8-10] how to identify hidden opportunities in drug repurposing projects, [11-13] or how to support the difficult but promising design of multitarget drugs [14-16]. Despite other rationale for target fishing presented here, additional information on potential off-target activity or selectivity of compounds from a starting library is a welcome side-product.
Computational target prediction methods published to date [13,17] can be classified as ligand-based, network-based, sideeffect- based, or protein-structure-based depending on the data used [18]. Ligand-based methods connect similarity measures with binding profiles for similar compounds in order to predict potential targets. Network-based methods incorporate the knowledge about ligand and target interactions, which are then represented as networks. Side-effect-based approaches utilize the information about off-target activities of similar drugs.
Potential targets can also be predicted by protein structure-based methods including docking, protein-ligand interactions or protein binding site comparisons, but this is a tedious manual procedure solely based on profound expert knowledge.
Quite new is the inverse approach-to create ligand bioactivity fingerprints encoding the hit status of compounds from HTS campaigns [19,20]. In combination with conventional ligand fingerprints those allow to identify chemically similar ligands that should have similar bioactivity profiles.
Ligand-based methods are fast and easy to use, but they are limited to search spaces of highly similar compounds. To a certain extent, they are able to extrapolate into new chemical space via scaffold hopping.
Docking, on the other hand, is dependent on the availability of protein crystal structures. For about half of the targets relevant to pharmaceutical research there are no crystal structures available. Docking, in principle, can identify new chemical matter, but it is challenging with respect to protein pre-processing and ligand ranking [18].
Pharmacophore methods, finally, are somewhere in between. To some extent they can extrapolate by scaffold or substituent hopping. On the other hand, pharmacophore methods often provide the user with an overwhelming manifold of hypotheses that without detailed SAR knowledge cannot be separated into meaningful and chance models.
Weighting the pros and cons of the former concepts we decided for a hybrid approach. We filter down the published - highly incomplete and sparsely populated - pharmacological universe by fast ligand-based methods to a manageable subset. We then process a user-selected subset of the ligand hit sets related to specific targets by docking. Our approach is as far as possible automated for efficient identification of potential biological targets with co-crystal structures. The general process starts with multiple automated ligand-based similarity searches in the ChEMBL [21] database, which contains chemical structures of small molecules with their associated biological test results and targets. Consequently, grouping of hits based on biological target, extraction of structures from Protein Data Bank [22] via the accession codes and automated docking simulations are performed.
The approach is novel in the way how multiple computational methods are combined in an efficient process, providing the computational chemist with a holistic picture of potential hits based on the available knowledge. It is implemented in a way to automate the tedious manual work, to provide an expert with the capability to interact with results and to allow him to concentrate on decision-making.
Despite a high degree of automation of this process, the crucial step will always be the final one, where the real value is generated by the modeling expert, who will make decisions based on visual inspection and his experience in order to adjust the combinatorial library to selected target(s) by adding, replacing or removing chemical substituents, or exchanging a scaffold. As a result, one or more novel targeted libraries can be designed.
By our approach we will lose all those targets our library would show some activity on but where the published ligands are too dissimilar in 2D metrics. A part of those targets could be “rescued” by direct docking into the complete crystallized target space, but even then we would still miss some targets due to the shortcomings of rigid receptor docking.
We do not aim for the identification of a complete targetome for our library, but for the identification of targets that fit into the pathways of our medical indications. We will therefore not aim for the highest-ranked target, but for the one best fitting to our project portfolio.
It is also important to understand that we do not describe a process of automated ligand- and target-based virtual screening. Instead, the similarity searches are applied as a coarse filter to identify targets from which the expert selects targets of interest. Docking is applied to confirm target fit based on pose consistency between cocrystallized ligand, ChEMBL hits and docked library compounds and has to be seen as a sharper filter to finally identify the most appropriate target for our library. In this paper we present a concept and a first implementation of the process that can be easily adjusted to individual needs, like adding a corporate database of chemical structures and biological data, extending the range of similarity search methods, exchanging protein preparation and docking method or adding automated pharmacophore modeling. Though implemented in a commercial software solution, the described protocol can be also realized using other tools and software.
Methods and Process Description
The basic concept of our target-fishing approach relies on the “similarity principle”, [23] according to which similar molecules exert similar biological activities. Therefore, a combinatorial library in its whole, its subsets or individual compounds, that are similar to known actives, should be able to point at targets of interest. Promising targets, which were identified indirectly using ligand similarity, are then selected for further investigation via automated docking. Conceptually, this resembles the process of experimental target validation using chemical probes.
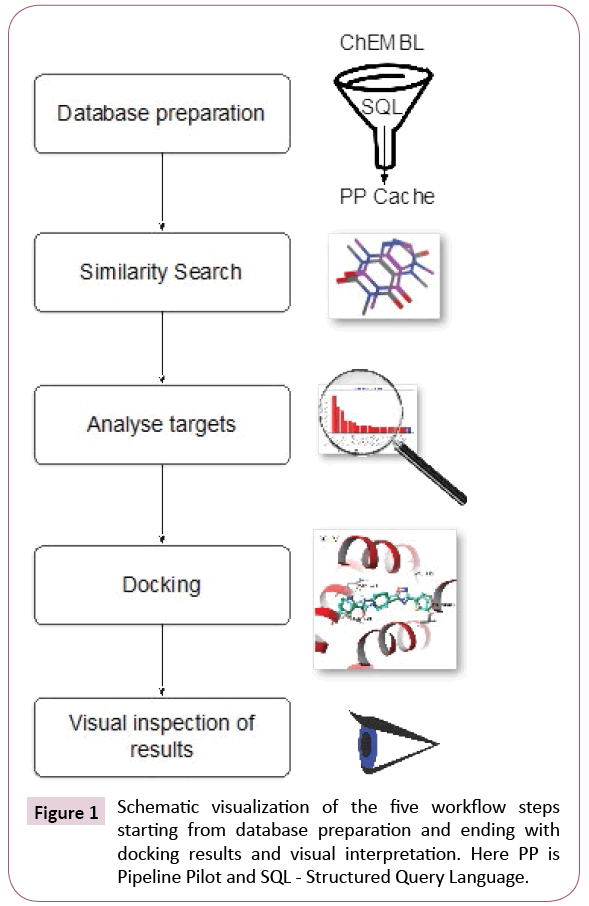
The automated protocol constructed and executed using the workflow software Pipeline Pilot [24] can be summarized into four steps, namely, database preparation, similarity search, analysis and docking, as it is shown schematically in Figure 1. In a fifth step, the computational chemists will visually inspect results and draw informed decisions.
Database preparation
From the broad range of data from the scientific literature, including biological activities for drug-like bioactive compounds as available in the public database ChEMBL [25], information about chemical structures, identifiers, assays and targets is extracted and saved into the appropriate file formats for the similarity searches in step 2. (The data in this work were based on ChEMBL version 14 (release from July 2012) comprising almost 14 million experimental results for about 1.9 million compounds, whereas the current release 23 from May 2017 contains around 2.1 million compounds). The database structure of ChEMBL consists of about 50 tables, which are mapped by primary keys and contain information about compound, source, drug properties, experimental data, target, mechanism of binding, etc. In order to access the most important entity types from the database, SQL queries were constructed and implemented in Pipeline Pilot to extract the data about compounds, targets, assays and activities, as well as adjustable filters for parameters like organism, activity type, activity threshold and confidence score.
Further investigation of the ChEMBL database revealed that there are more than 3000 different activity types measured in hundreds of different units. Among them the top-represented activity types, which were used in our study, are potency, EC50, IC50, inhibition, Ki. Moreover, grouping of compounds by organisms revealed 1621 species on which they were tested. Thus, we implemented a number of default filters for the most represented activity types (IC50, EC50, Ki, Kd), units (M, nM, μM, mM) and organisms (human, mouse and rat) as well as for the activity threshold (10 μM). Those filters can be easily set via Pipeline Pilot protocol checkboxes and variables.
To ensure as much as possible that targets are assigned to correct assays, only records with ChEMBL confidence score higher than 7 were selected. The confidence score is assigned during the manual curation process by the data extractors and reflects assay-target relationships. It ranges from 0 to 9, where 0 means uncurated data and 9 equals to high degree of confidence.
The application of above-mentioned filters reduced the amount of ChEMBL entries from 12.3 to 3.8 million, which represents 764,419 unique registered molecules. The compounds and the information about targets and assays were saved into separate files. Thus, all additional information was joined to compounds after the similarity search. We apply a predefined hierarchical file structure for the purposes of documentation and to facilitate further re-analyses and follow-up studies. Finally, the extracted ChEMBL data were converted into the appropriate structure formats required for the chosen similarity methods as described in the next step.
Similarity search
For each compound of a combinatorial library ligand-based virtual screens against database compounds are performed. Multiple methodologies are applied to make maximum use of different similarity measures. Final hit lists are combined by MAX-rank voting as described by Baber et al. [26] and Whittle et al. [27].
In this work we implemented three approaches, namely (i) atom-based circular fingerprints ECFP4, (ii) non-linear Feature Tree descriptor FTrees and (iii) DBTOP topomer search similarities. Each of them represents structural and pharmacophoric features in a different and complementary way.
The extended connectivity fingerprints ECFP4 describe the presence or absence of overlapping particular substructures [28]. The number 4 in the name corresponds to the effective diameter of the largest feature, thus the largest possible fragment has a width of 4 bonds. The Tanimoto coefficient is used as distance metric for scoring.
DBTOP from Certara is a 3D similarity search where molecular structures are compared as sets of fragments (so-called topomers), which are characterized by CoMFA-like steric shape and pharmacophoric features [29]. One single rule-based conformation is generated for each fragment and oriented by open valence bond, while the rest is oriented again using a rulebased scheme. Aligned fragments are then compared by their fields until the minimum topomeric difference between two molecules is identified.
The BioSolveIT FTrees method calculates the feature tree descriptor, which represents hydrophobic fragments and functional groups of the molecule and the way these groups are linked together [30]. The descriptors of two molecules are then compared to each other.
ECFP4 and FTrees are available as Pipeline Pilot components, whereas DBTOP was run from the command line using Pipeline Pilot “Run on Server” component. Since we aim for target fishing and idea generation, we accept low overall ligand similarities and therefore limit hit lists of the individual searches by the maximum numbers of hits and not by similarity thresholds.
The implemented Pipeline Pilot protocol allows a user to select similarity search methods via checkboxes and to set individual parameters for similarity threshold or number of top-hits to save. It automatically combines results of similarity searches and reports hits, their similarity scores as well as targets, activity and assay data.
Analysis and selection
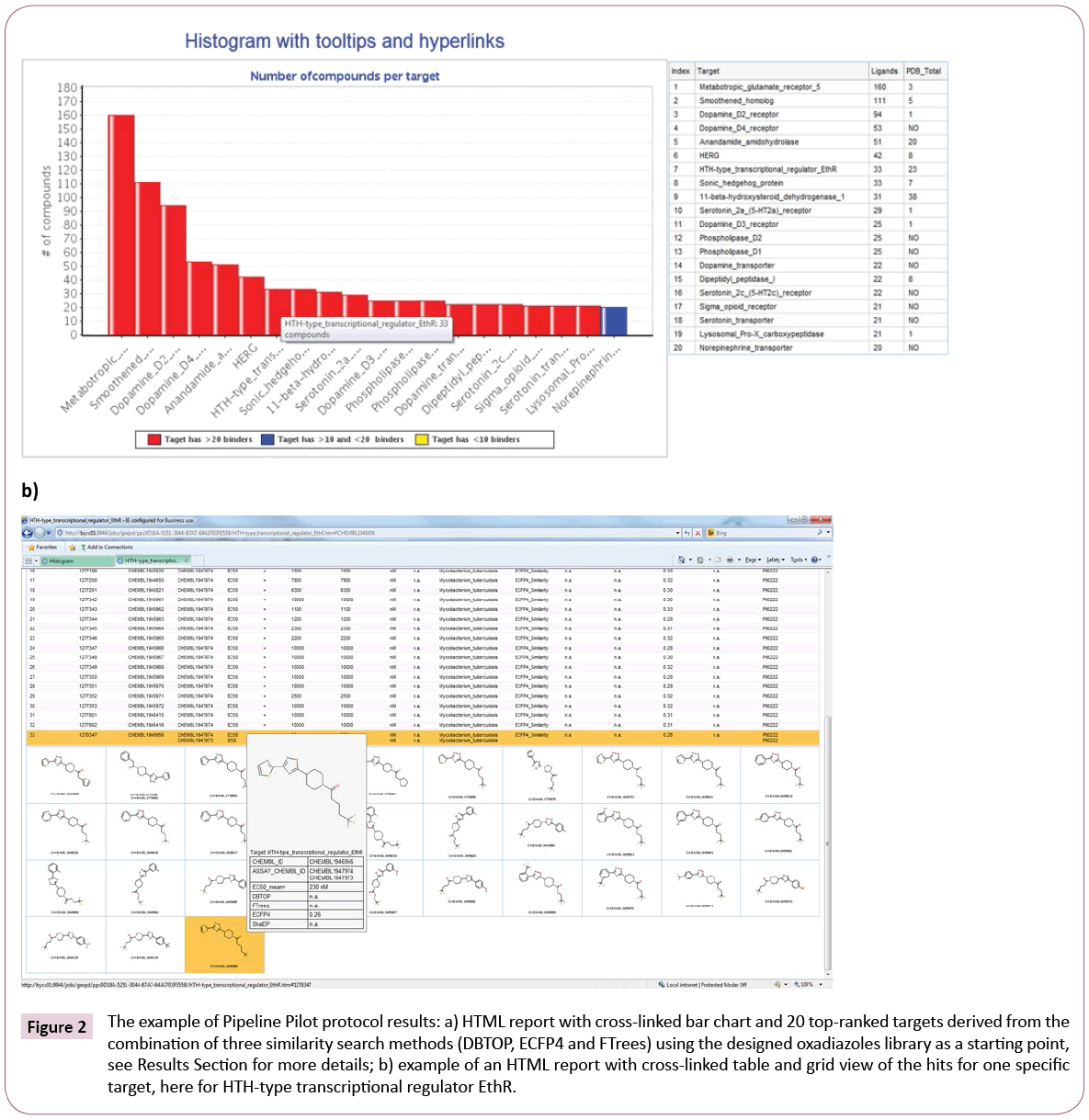
Hits are grouped based on the targets against which they show activity. The results are presented as Pipeline Pilot HTML report comprised of an interactive bar chart, representing top targets and numbers of hits per target (Figure 2a).
Figure 2: The example of Pipeline Pilot protocol results: a) HTML report with cross-linked bar chart and 20 top-ranked targets derived from the combination of three similarity search methods (DBTOP, ECFP4 and FTrees) using the designed oxadiazoles library as a starting point, see Results Section for more details; b) example of an HTML report with cross-linked table and grid view of the hits for one specific target, here for HTH-type transcriptional regulator EthR.
The ranking order implemented here is disputable, since currently targets are sorted by number of hits identified, which yields a certain bias towards targets with higher numbers of congeneric compounds reported. Since the rank score bears a certain risk of missing interesting targets with small hit clusters, the user is able to set a threshold for the number of targets retrieved. Up to now, for each input library we were able to identify a set of interesting targets. Nevertheless, alternate scoring schemes taking into account, for instance, overall numbers of compounds tested, activity ranges, and numbers of congeneric series will be evaluated.
For convenient overview the bar chart is equipped with tooltips and hyperlinks, showing the full target name and hit counts for the different similarity measures. Since we are solely interested in targets with crystal structures, information about protein structure availability is also retrieved from RCSB Protein Data Bank [22] and summarized in the table next to the bar chart, see Figure 2a.
Furthermore, a click on any bar of the chart executes a Pipeline Pilot sub-protocol, which provides a second HTML report (Figure 2b) containing table and attached structure grid view with detailed information about the hits, e.g., chemical structure, activity data, assay results or species on which they were tested. Moreover, the table area and the grid view are cross-linked and possess tooltips containing chemical structure and detailed assay information. This gives the user a quick overview of a certain target and its compounds as well as assists with further target selection. The desired targets can be preselected for docking in the next step using checkboxes.
Docking
Automated docking of library compounds, ChEMBL hits and cocrystallized ligand into the selected targets is performed. All available PDB structures for user-selected targets are downloaded by the workflow, i.e., often multiple crystal structures per target. For instance, the amount of structures deposited in PDB for cyclindependent kinase 2 is more than 300. This poses a question how to prioritize the crystal structures for docking in an automated way. One quality criterion for a crystal structure, which can be easily accessed, is its resolution. On the other hand, docking may be still not successful, when it is done into a wrong protein conformation. Since residues of apo-structure (without bound ligand) may occupy parts of the binding pocket, we decided to limit our docking to holo-structures (ligand-bound). Furthermore, the presence of a ligand simplifies automated grid generation. Thus, top N holo-structures with the best resolution are selected for each target, where N is a number specified by the user. In case of multiple chains, always chain A is saved for each structure in order to simplify structural alignment. Alternative selection schemes could include target selection by ligand similarity or pocket shape diversity.
Ligand preparation was done in two steps. First, protonation states at pH 7.4 for co-crystallized ligand, ChEMBL hits and library compounds were calculated using the pKa module co-developed by Bayer and SimulationPlus [31] and implemented as Pipeline Pilot component “ADMET predictor” [32], while ring conformers, tautomers and stereoisomers were generated using Schrödinger LigPrep utility, release 9.8.
An automatic docking procedure was applied using the Schrödinger script XGlide.py (version 3.7; v45017). The script performs automatic protein alignment and preparation, grid generation, re-docking of crystal structure ligands as well as docking of other compounds (here, library compounds and ChEMBL hits). For each selected target a separate directory is created containing subdirectories for crystal structures, prepared ligands and docking results. The script is executed from the command line using Pipeline Pilot component “Run on Server”. The following docking parameters were applied: protein alignment, preparation and grid generation were turned on; ligand preparation was set to false, Glide standard precision (SP) was selected as the scoring function. The results for each target were saved as pose viewer files, which at the end are copied into one folder for the analysis.
Inspection
The final step in the process is by intention not automatic, and probably can never be. The computational chemist loads docking poses for targets of interest for visualization and analysis. In the first step he inspects the quality of re-docking of co-crystallized ligands and identifies commonalities and differences in the binding modes to individual crystal structures of each target. In the second step, he inspects docking of ChEMBL hits to verify the interaction hot spots. Third, he analyzes the library compounds with good and bad docking scores and judges the plausibility of the binding modes obtained. Finally, he will either consider biological testing of library compounds on targets of interest, or modifying the library proposals in order to optimize their interactions to a certain target, or generation of a completely new library proposal.
Results
Validation of similarity search process
Of the three purely automated technical steps, namely database preparation, similarity search and docking including grid preparation, the most critical one for the overall performance is the identification of targets via the similarity searches. We therefore preformed a retrospective study in ChEMBL to test for the performance of finding targets via searches with libraries known to be active on those targets.
For this, we extracted ChEMBL data for compounds tested on all species with reported IC50, EC50, Ki, Kd and activity units of nM or μM. No activity threshold filter was set. The applied filters reduced the number of ChEMBL entries to 851,915 which constitute 327,520 unique molecules and 17568 different DOC_ IDs [Fussnote einfügen: DOC_ID, TARGET_ID, MOLECULE_ID all have the same identifier name CHEMBL_ID in different tables of the ChEML database]. From those, 633 sets based on identical DOC_ID were derived containing between 100 and 150 molecules each, representing our chemical libraries. This is justified by the fact that compounds from one publication normally more or less represents a congeneric series. The 633 sets are connected to 264 different TARGET_IDs. We finally selected 22 DOC_ID sets which share their TARGET ID with 5 to 7 other documents (the distribution runs between 1 and 13 different documents per TARGET_ID).
This setup allows us to perform - using the compounds from one document - a “library-based” similarity search. By those similarity searches we should then be able to re-find the target the search library is known to be active on, only based on similarity of the library compounds to the compounds in other documents on the target.
Detailed results are provided in Table S1 in ESI. The median numbers of documents identified are 45 for the combined search and 10, 43, and 7 for ECFP-4, DBTOP and Ftrees, respectively.1
We are thus always able to identify the targets of the library compounds even though the median similarities to the ChEMBL compounds are as expected quite low with 0.33 for ECFP-4, 139 for DBTOP and 0.88 for Ftrees. With two exceptions all targets were identified by all three methods. Tyrosine-protein_kinase_ SYK (CHEMBL2599) was not found by ECFP-4 and Ftrees and Cytochrome_P450_2D6 (CHEMBL289) by ECFP-4.
Thus, we are consistently able to identify the target we were looking for, but not always at rank 1. Nevertheless, mean ranks of the test targets are 3.5 for ECFP-4, 8.7 for DBTOP, 5.8 for Ftrees and 1.4 for the consensus rank, which always ranks the search target rank 1 or 2.
The hit rate and especially the ranking of the search targets is even better than the expected outcome, i.e., that the similarity searches are performed to identify a shortlist of targets for selection by the expert, not to identify the rank 1 targets.
Process application examples
The process described in Methods and Process Description was developed to identify potential targets for existing chemistrydriven combinatorial library proposals and to modify the proposals in a way that they can directly contribute to early projects at Bayer Pharmaceuticals Global Drug Discovery. The process is applied to in-house libraries that are proprietary and cannot be disclosed here. Therefore, we had to design a proof of concept case study for this publication. The downside of this approach is that we are not able to present experimental data for our prospective library proposals (the starting library or the derivatives for the targets we hit). As a starting point we chose a publication from the Journal of Medicinal Chemistry from 2012 which describes structurebased drug design for a series of potent 1,2,4-oxadiazoles, which target M. tuberculosis transcriptional repressor EthR (see Figure 3a for examples) [33]. We designed a combinatorial library, that is similar but distinct to the published compounds from ChEMBL, with the aim to demonstrate that the developed methodology is able (i) to recover the compounds from the publication and to show that EthR protein can be identified among the top targets, (ii) to identify potential new targets for our example library, (iii) to provide examples of target-fishing-based library modifications and (iv) to provide examples of the short-comings of such a fully automatic approach and to highlight the importance of expert interaction.
In particular, we introduced three changes to our library with respect to the library from the publication. First, we modified the piperidine ring to a cyclo-hexyl, i.e., shifted the nitrogen by one position. Second, we replaced the aliphatic lipophilic side chain by various R2 groups of different size, polarity and charge state, connected via nitrogen or amide bonds. Third, we introduced alternative lipophilic R1 groups at the only point of variation from the published library. Core definitions and examples for the publication and the library compounds are shown in Figures 3a and 3b, respectively.
Example of database preparation: Step 1 of the workflow is to search for similar compounds and their associated targets using the designed library as a reference. For now, ChEMBL data were extracted for compounds tested on all species with reported IC50, EC50, Ki, Kd and activity units of nM or μM. No activity threshold filter was set. The applied filters reduced the number of ChEMBL entries to 851,915 which constitute 327,520 unique molecules.
Example of similarity search: In step 2, similarity searches are performed. We used all three currently implemented methods, namely DBTOP, ECFP4 and FTrees as described in Methods. 400 highest rank hits were saved for each metric, and additionally a consensus rank was calculated. The diagram in Figure 2a gives the list of the top 20 targets, associated with the results of the similarity searches, as interactive bar chart. Table S1 of Supporting Information provides more detailed information about target ranks according to the three similarity search methods and consensus rank; additionally, it lists the numbers of identified hits and PDB structures for each target. The targets in Table S1 are sorted by descending number of ligands identified by ECFP4 similarity search. As mentioned earlier, the implemented ranking by number of hits per target may be biased towards the targets with large congeneric series.
Example of analysis: Step 3 is the first of two expert intervention steps. Target selection could be done automatically based on their ranks, but manual selection will allow to concentrate on targets relevant in the context of a company’s research portfolio.
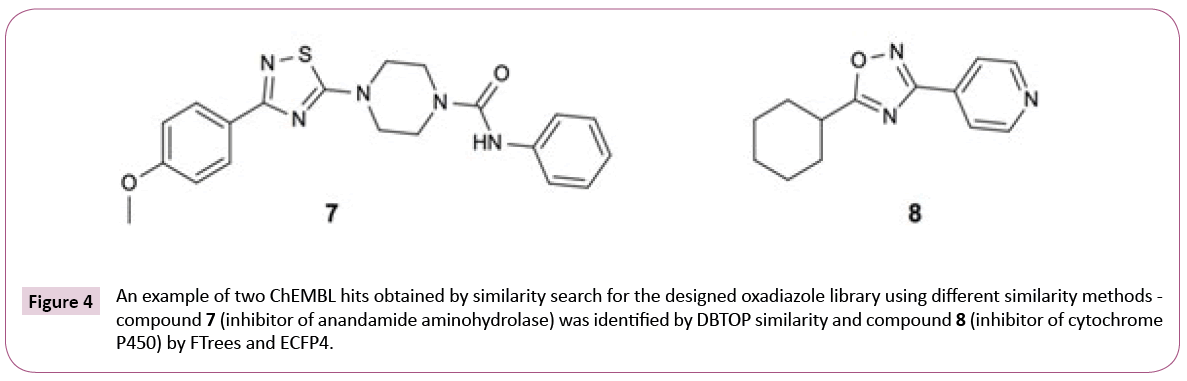
The transcriptional repressor EthR was ranked number 7 by the consensus score, which combines the results of the three similarity search methods. The scoring according to ECFP4 method ranked EthR on position three. ECFP4 was able to identify all 33 compounds from the publication, whereas FTrees found only 2 and DBTOP none, underlining the necessity to apply multiple ligand-based search methods to obtain the complete picture. DBTOP is based on steric and pharmacophoric fields of the whole molecule and therefore is more susceptible to larger size differences between query and database molecules than FTrees, which abstracts the molecular fragments into pharmacophoric representations, or ECFP4 circular fingerprints, where the hits are dominated by occurrences of fragment features. Depending on library, contributions of different methods will differ. Some inhouse library screens, for instance, were dominated by DBTOP hits. It is a priori not obvious which similarity metric will dominate in the consensus hit list.
Figure 4 shows the example hits identified by different similarity search methods to underline this assumption. It is worth to note that similarity scores (see Table S2 of Supporting Information) as expected are quite low, pointing out that chemical modification of the library compounds guided by the final docking step might be needed.
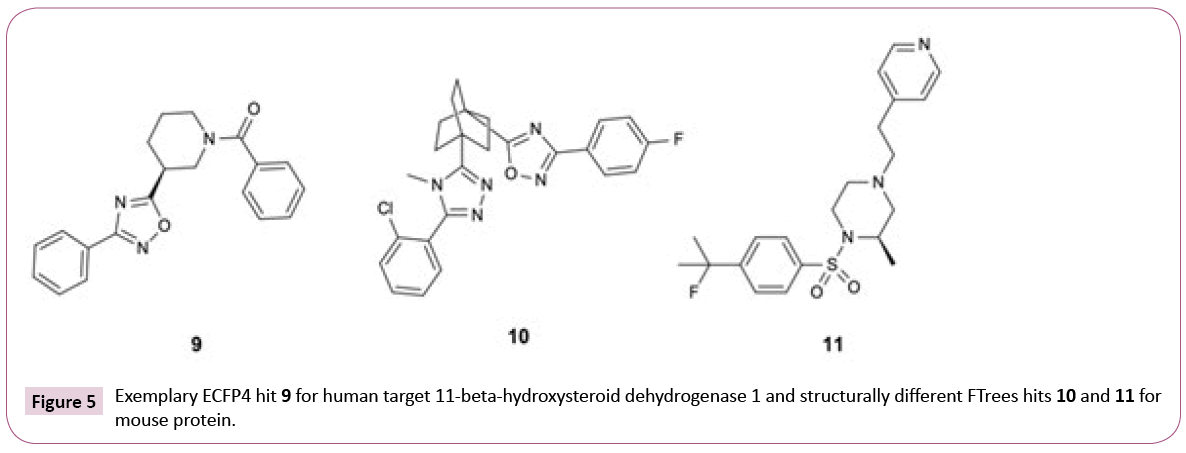
As expected, the numbers of hits for the different search methods differ. But in addition, also the numbers of PDB structures retrieved differ. For instance, 33 PDB structures of 11-beta-hydroxysteroid dehydrogenase 1 are found using only ECFP4 (see Table S1), whereas the combination of three similarity methods retrieved 38 PDB entries. The reason for this lies in the fact that all ECFP4 hits are annotated with UniProt [34] identifier P28845 (human) whereas the combination of ECFP4 and FTrees resulted in hits, which were tested on human and mouse 11-beta-hydroxysteroid dehydrogenase 1 (UniProt identifiers P28845 and P50172, respectively). While the human sequence shares 79% identity to the mouse orthologue, there is high level of conservation of amino acids in the binding site. All ECFP4 hits share the same oxadiazole motif while FTrees identified two additional motifs (Figure 5). Again, it is strongly emphasized that it is advantageous to employ multiple ligand similarity metrics.
Our proof-of-concept target EthR is rank seven by consensus score and rank three by ECFP4 similarity search. In the following we will analyze the two top-ranked targets in more detail (see also Table S1), together with our target of interest, EthR (which would resemble the real-life situation with some targets in the list not being relevant for the current portfolio.
Example of docking: For step four we selected the two top-ranked targets for docking, namely, top-ranked target metabotropic glutamate receptor 5, second-ranked receptor smoothened homolog, and our proof-of-concept target HTHtype transcriptional regulator EthR which is ranked seventh. The docking of our library compounds, ChEMBL hits and cocrystallized ligands was performed using the fully automated XGlide procedure as described in Methods. A maximum of 2 crystal structures per target were retrieved automatically. We had to extend the set by one more structure in the case of EthR, as described in the following.
Our decision objective for target fit is correct re-docking of the co-crystallized ligand, consistent docking of the ChEMBL hits and finally consistent docking of the library compounds or similarly decorated subsets thereof. We provide docking scores as a means of further confirmation of consistent placement, but not as a filter or design criterion per se. High docking scores are a strong hint for important interactions to the target matched, whereas low scores are not always correlated to weak binding interactions.
Helix-Turn-Helix-type (HTH-type) transcriptional regulator EthR
Currently there are 23 protein structure entries in RSCB protein data bank based on UniProt ID accession code P9WMC1 (Mycobacterium tuberculosis). Since the number of structures for docking is actually a compromise between expected information gain and effort, two structures for docking were automatically selected from the 17 holo-structures available, based on crystal structure resolution. By default, we process two different crystal structures since modelling experience tells that using multiple target structures for rigid docking reduces the risk of missing important target information. We later added one additional structure, namely 3O8H, due to its different pocket shape and ligand-binding mode.
The hits found by ECFP4 are both agonists and antagonists with best EC50 of 60 nM and IC50 of 400 nM, respectively, i.e., highly active compounds.
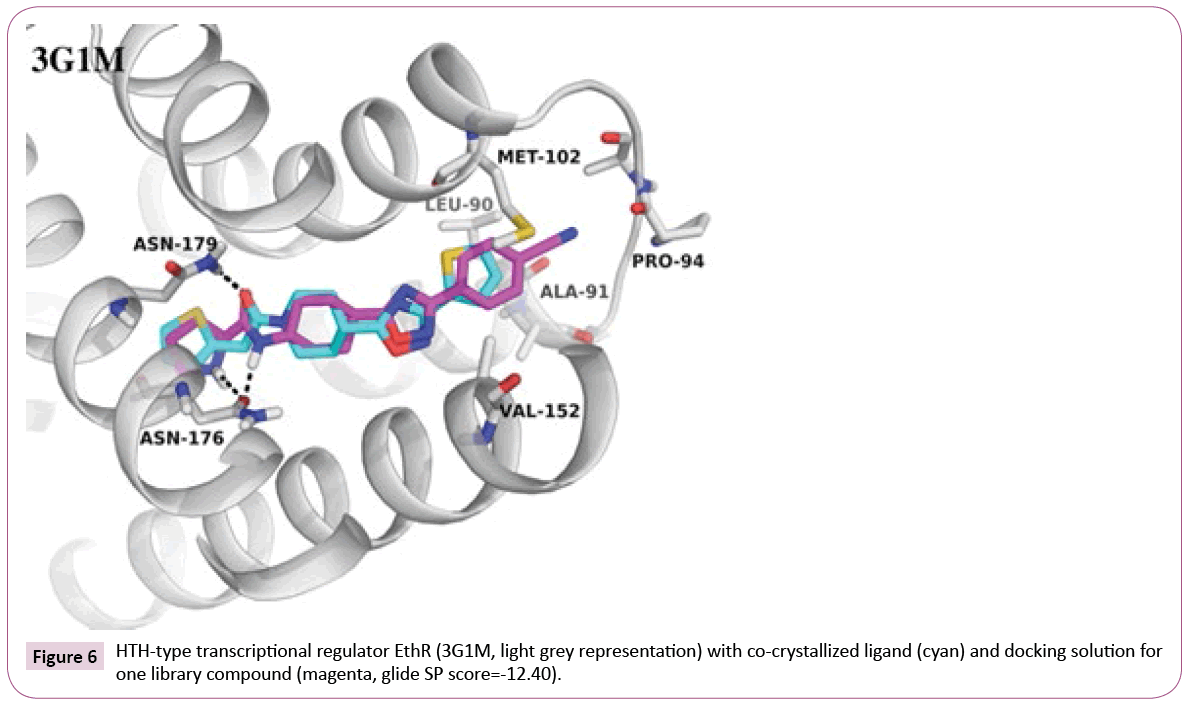
G1M: An example where library fits well into the target: Docking of the library compounds into the first crystal structure 3G1M with a resolution of 1.7 Å yields in high docking scores and poses comparable to the co-crystallized ligand (IC50 of 500 nM, retrieved from PDB Bind [35]). An additional hydrogen bond to Asn176 can be observed between EthR and some of library compounds containing tertiary amine or amide linker attached to the oxadiazole-cyclohexane core (an example can be seen in Figure 6). In contrast, the co-crystallized ligand, which has an oxadiazole-piperidine scaffold, is missing a hydrogen bond donor at this position. Moreover, the analysis of the binding pocket around the ligand can provide further suggestions for compound modifications, e.g., for extended interactions into the hydrophobic pocket formed by Met102, Val152, Leu90.
3Q0W: differences in protein conformation and incomplete binding site setup: In contrast, docking into the second EthR structure (co-crystallized ligand has Ki of 400nM [35]) led to low-scored poses for our library compounds. It turned out that a cocrystallized glycerol molecule, that had not been removed by the automated protein preparation, was situated deep in the binding site, establishing hydrogen bond to Asn176 and blocking ligand entry.
After its removal, docking of all compounds was possible. Nevertheless, the poses are still quite inconsistent. The amide moiety for about half of the poses is located deep in the pocket and makes hydrogen bonds to Asn176 and Asn179, analogously to the 3G1M dockings shown in Figure 6, and for the other half it points out of the pocket. Such differences can be explained by conformational flexibility of the protein, which can be seen in comparison of the two EthR crystal structures (PDB codes 3G1M and 3Q0W, the superimposition is shown in Figure S1, see Supporting Information). Slight but pronounced differences can be observed at the loop region (residues Asn93-Asp98), where the flip of Pro94 is accompanied by narrowing the entry channel, which sterically hinders the placement of substituents towards this loop in 3G1M.
3O8H: Alternate binding mode: Our library was intentionally designed to be chemically similar to the EthR inhibitor BDM41906 [33] (PDB ID: 3SFI). 3G1M and 3SFI have the same overall shape, the library compounds dock consistently into both pockets (results are not shown).
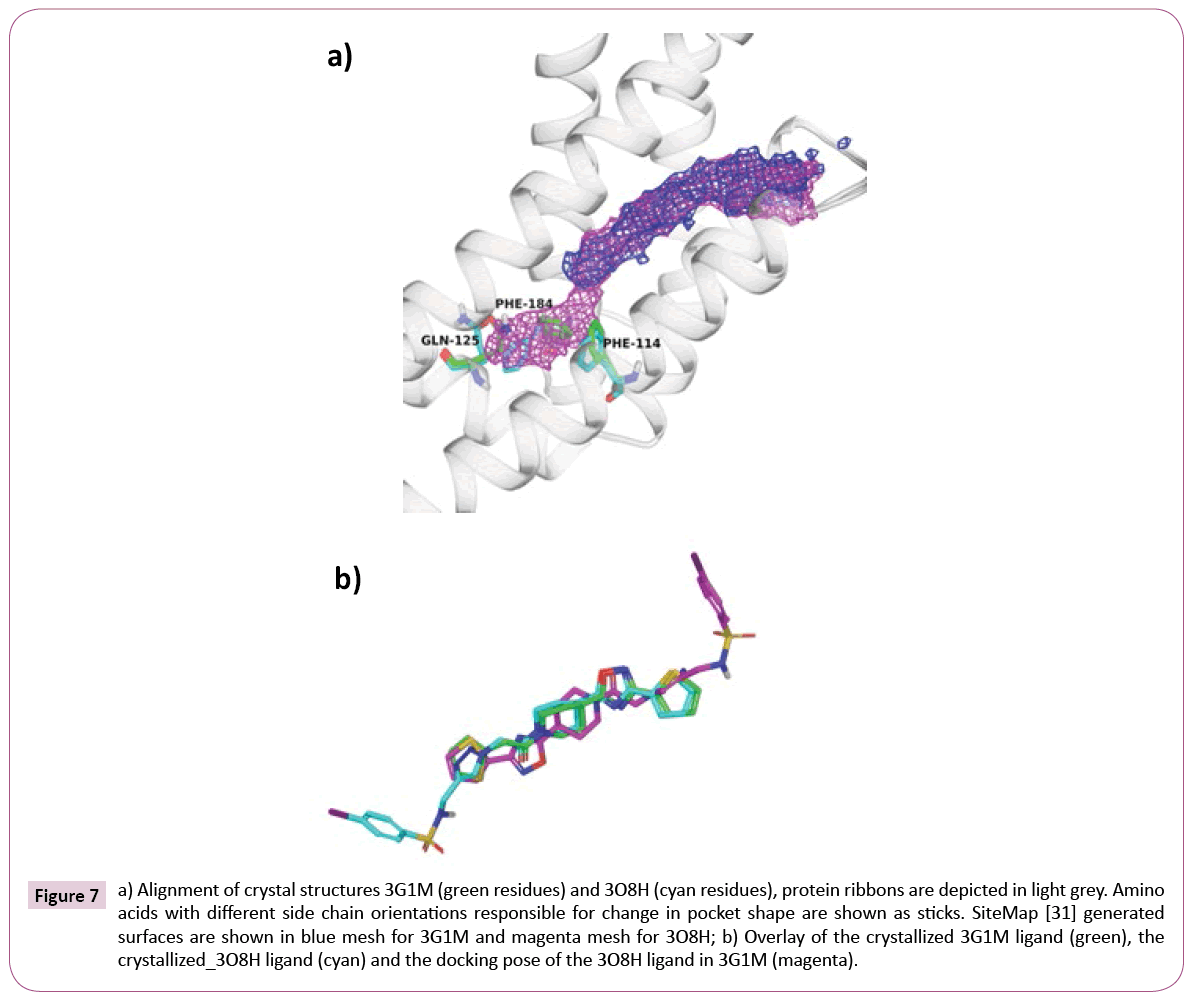
Nevertheless, closer inspection of EthR structures revealed a second set of crystal structures with considerably larger binding pocket. Such pocket enlargement is mainly caused by the flip of side chains of Thr121, Gln125, Trp138 and Phe184 (see Table S3 for comparison of available EthR crystal structures).
Figure 7a shows the alignment of 3G1M and 3O8H along with interaction volumes generated by SiteMap [36]. As expected, cross-docking of the 3O8H ligand (IC50=580 nM) into the rigid 3G1M receptor, without taking into account any induced fit effect, yields a completely different and wrong binding mode, where the aromatic sulfonamide is pointing out of the pocket (see Figure 7b).
Figure 7: a) Alignment of crystal structures 3G1M (green residues) and 3O8H (cyan residues), protein ribbons are depicted in light grey. Amino acids with different side chain orientations responsible for change in pocket shape are shown as sticks. SiteMap [31] generated surfaces are shown in blue mesh for 3G1M and magenta mesh for 3O8H; b) Overlay of the crystallized 3G1M ligand (green), the crystallized_3O8H ligand (cyan) and the docking pose of the 3O8H ligand in 3G1M (magenta).
About two thirds of the library members dock consistently to BDM41906. About one third, due to the pronounced pocket differences, dock inconsistently. Library members from both sets ignore the additional cavity available in 3O8H.
In summary, we were in fact able to identify EthR as a potentially interesting target based on ligand similarity and docking results for our designed oxadiazole library. We have also demonstrated, that further optimization strategy largely depends on the choice of EthR crystal structure, since the pocket residues are the subject of conformational changes. Based on the docking results from both pocket shapes, we gained worthwhile additional information about flexible and rigid subpockets and key interaction features. If it were for our library extension campaign, we would now, based on the target information, slightly optimize the decoration of the initial library and additionally design a second library that targets the deep cavity available in 3O8H. We would cross-check the design for IP space and if necessary iteratively adjust to create novelty.
Metabotropic glutamate receptor 5
The highest ranked target according to ECFP4, the metabotropic glutamate receptor 5, is a class C G-protein-coupled receptor responding to the neurotransmitter glutamate. There is only one holo structure (PDB ID 4OO9) identified in PDB for the transmembrane ligand-binding domain, since earlier structural studies had been restricted to the amino-terminal extracellular domain, providing little understanding of the membranespanning signal transduction domain. 4OO9 is co-crystallized in complex with the negative allosteric modulator, mavoglurant.
The similarity searches for the library compounds identified in total 160 agonists and antagonists of the metabotropic glutamate receptor 5 using consensus scoring, with best affinity values of EC50=5 nM, IC50=130 nM, and Ki=150 nM. The ECFP4 method ranked this target at the top position, while FTrees ranked it at the position three with 149 and 25 inhibitors being identified, respectively. There were no metabotropic glutamate receptor 5 inhibitors among top 25 targets identified by DBTOP method. The hits represent different structural clusters such as piperidine-amides, piperidine-sulfonamides and spiro-hexyl-4,5- dihydrooxazoles (see Figure 8 for examples).
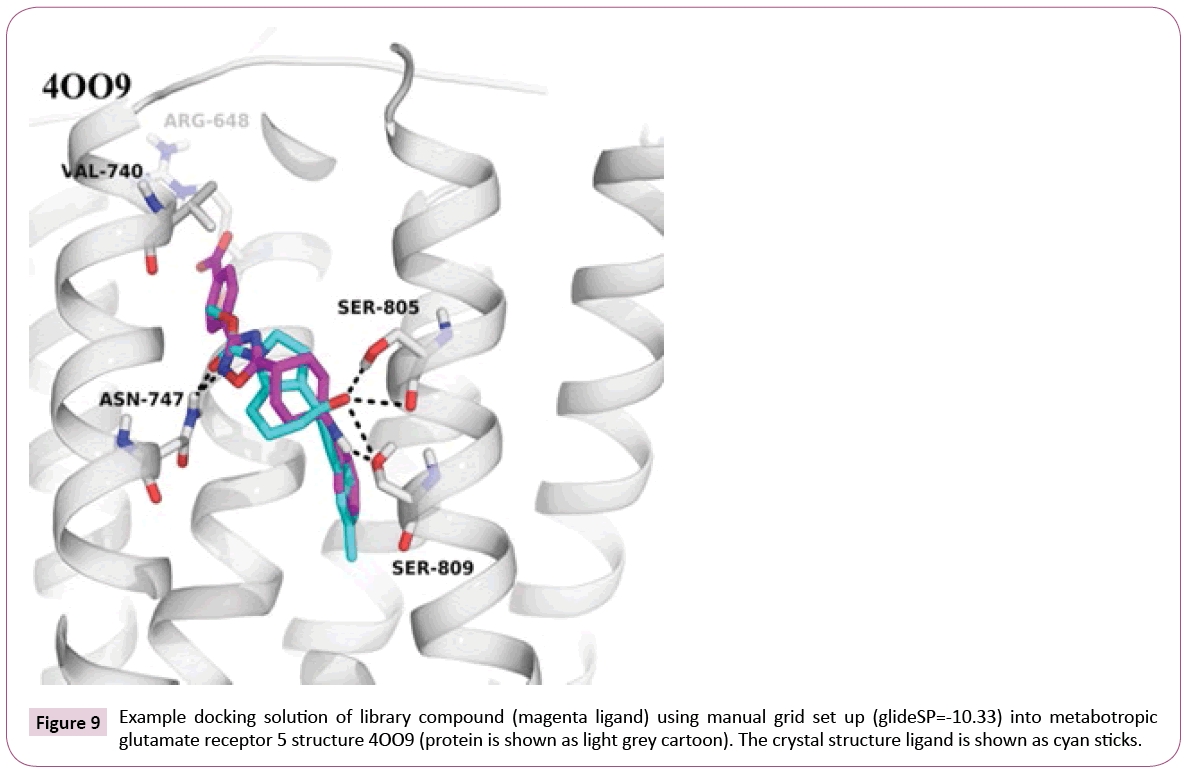
4OO9: Failure of the automatic procedure: All steps of the automatic workflow technically proceeded well and compounds were successfully docked. However, a closer look at the crystal structure 4OO9 revealed that during automated protein preparation and docking, the docking grid was positioned around a co-crystallized small organic molecule coming from the experimental conditions, namely oleic acid, and not around the allosteric modulator mavoglurant [37]. Thus, the docking was performed into the wrong pocket (see Figure S2 of Supporting Information).
The example of 4OO9 shows that the fully automated docking procedure has its drawbacks. Protein preparation and docking setup require user inspection and in certain cases manual correction. The effort nevertheless is acceptable, since the expert should be knowledgeable of the target in order to understand and to judge the observed ligand interactions. On the other hand, one could implement a mechanism to retrieve information about the actual ligand and its binding mode, and use it during the protein preparation step.
Docking into the manually prepared binding site reveals that our library compounds exhibit numerous interactions to the receptor similar to the crystal structure ligand, e.g., hydrogen bonds to Asn- 747 and Ser-809, and extend their interactions deeper into the pocket lined out by Arg-648 and Val-740 (see Figure 9), which can be further analyzed to guide possible compound modifications.
Smoothened homolog
The Smoothened (SMO) receptor is a key signal transducer in the Hedgehog (Hh) signalling pathway. SMO is classified as a class F (frizzled) G-protein-coupled receptor (GPCR). It contains the conserved seven-transmembrane helical fold common to the class A GPCRs and an unusually complex arrangement of long extracellular loops stabilized by four disulphide bonds.
The similarity search for the library compounds identified overall 111 SMO inhibitors with a best IC50 of 16 nM, using consensus scoring. 103 hits arose from ECFP4 search and 20 hits from FTrees. All hits are piperidine-amides or piperidine-ureas, but again FTrees was able to detect more diverse compounds.
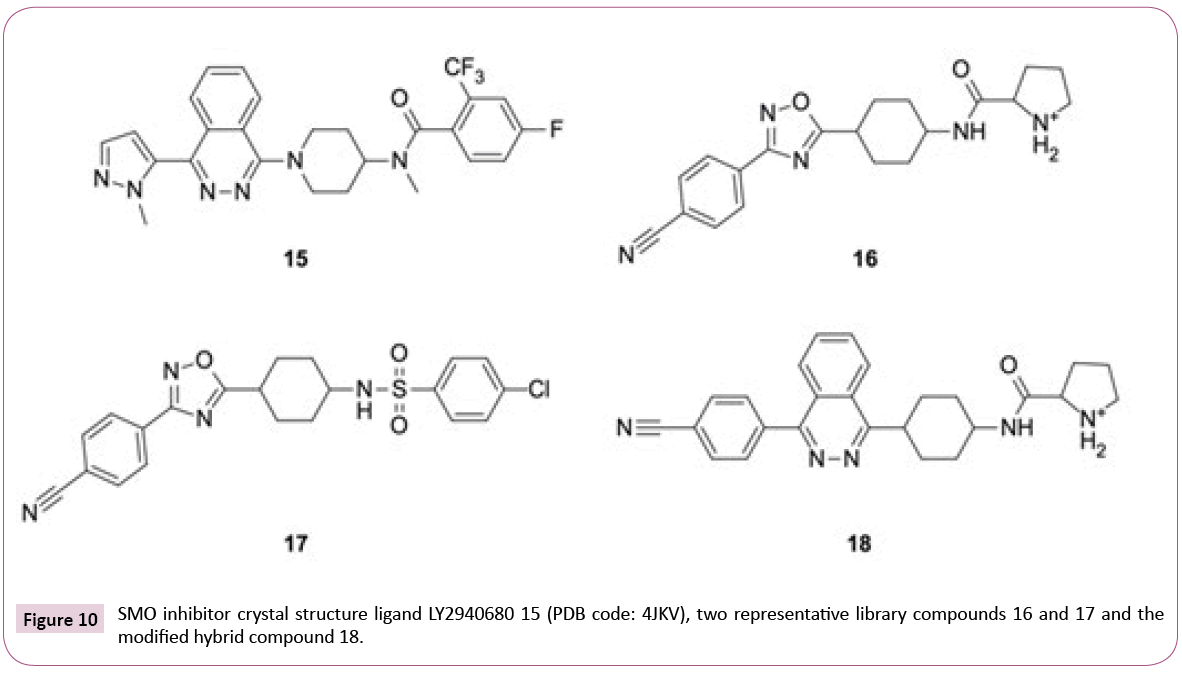
4JKV: Optimizing the library for the target: The 2.5 Å resolution crystal structure of the human SMO receptor contains the transmembrane domain together with antagonist LY2940680 15 (see Figures 10 and 11), which binds the extracellular end of the
Figure 11: Docking into the smoothened (SMO) receptor (4JKV: protein is shown as light grey cartoon). a) redocked LY2940680 15 (cyan sticks) and docking poses of two library compounds 16 and 17 (green and magenta); b) LY2940680 15 (cyan) and the docking pose of modified hybrid 18 (green sticks), the phtalazine cores of both compounds are well-aligned.
seven-transmembrane-helix bundle via extensive contacts to the loops. Redocking reproduced the binding mode with an RMSD of 0.52 Å.
Docking into Smoothened homolog resulted in many well-scored solutions for the ChEMBL hits and library compounds with consistent docking poses (see Figure 11 for examples).
Instead of the conserved hydrogen bond between the carbonyl group of ligand and Asn219, which is observed for most of the ChEMBL inhibitors and LY2940680, the library compounds form one or two (e.g., ligands with positively charged aliphatic ring like compound 16, see Figures 10 and 11) additional hydrogen bonds with the backbone carbonyl of Tyr394.
On the downside, most of the library compounds do not pi-stack to Phe484. Exceptions are, for instance, sulfonamides like 17 (see Figure 11). One of oxadiazole nitrogens of library compounds usually participates in hydrogen bonding to Arg400 analogous to the phtalazine nitrogens in the crystal structure [38], but none of the library compounds is able to fill the hydrophobic pocket occupied by the phtalazine core.
The observations about key interactions of LY2940680 15 and the ChEMBL compounds provide us ideas for possible library modifications in order to target SMO binding pocket optimally.
Figure 11b shows an example of hybrid compound 18 which has the oxadiazole replaced by phtalazine core while keeping the larger p-cyanophenyl and the positively charged pyrrolidinoamide. The Glide SP docking score for 18 (-12.39) is virtually identical to 15 (-12.66). Alternate hetero-bicycle replacements also fit well. Aromatic headgroups linked via sulfonamide or amide like in compound 17 (glideSP=-11.47), on the other hand, are able to participate in pi-stacking with Phe484. Overall, from the point of view of target interactions, there is a bunch of options, with similar but also better Lipinski properties than LY2940680 having a molecular weight of 512 Da and an AlogP of 4.56.
Conclusion
When planning for the extension of a compound library, one is confronted with a universe of synthesis options. The only limitations, thus, are one’s own creativity, lab and budget resources in order to transform the ideas into chemical libraries. Therefore, a rational concept to explore the options and pick the libraries with a certain probability to hit biological targets is desirable. Expert knowledge can guide the planning procedure. However, in such case the library design can be limited to the person’s experience around the projects he has worked on. Metrics like ligand efficiency allow to stay in an attractive property profile range but do not assist the selection of compounds amenable to target families of interest. Although drug-likeness or target class-likeness scores take into account overall similarity to known drugs or actives, substructure or global pharmacophore features are quite rough estimates of target family fit.
In this paper we therefore describe the productive implementation of a concept aiming, on one hand, to identify putative targets for a chemistry-driven library proposal and, on the other hand, to identify options for compound modifications in order to create new libraries better fitting to certain targets. This article reports on a designed virtual combinatorial library and hits identified by the workflow, as well as on library modification ideas without the desirable proof of synthesis and experimental testing.
The real in-house examples cannot be disclosed here, and the examples shown will not trigger any synthesis and testing at Bayer. Currently we are still not able to provide significant statistics about success rates of the described workflow due to its novelty and the long turn-around times for the process of library design, out-sourced chemical realization, registration and testing.
Our workflow aims to automate all tedious time-consuming technical steps and allow to concentrate on rational design. We always start with a chemistry-driven library carefully checked for novelty and end up with a proposal that again is checked for novelty as a part of the design procedure.
The workflow is divided into a set of protocols implemented in Pipeline Pilot that control the crucial steps, run automatically and require minimum user interaction which is productively used at Bayer Drug Discovery. The implementation described in this paper compares a virtual compound library to the chemical space represented in ChEMBL by multiple ligand-based similarity metrics, retrieves ligand and target information and presents the results in an intuitive representation to an expert, who then decides whether to proceed with ligand design for targets of interest. The protocol automatically retrieves PDB structures and sets up docking runs for the cocrystallized ligand, the ChEMBL compounds and library structures. The most timeconsuming step is, by design, the final one, i.e., the visual inspection by a computational chemist, who can further trigger library modifications or re-design. The system is easy to use and it is highly productive. The workflow is modular and can easily be extended to alternate database sources, similarity metrics, hit prioritization algorithms, or docking protocols. Due to its accessibility, the ChEMBL database was chosen as a source of biological and chemical information. However, incorporation of alternate data sources is obvious.
As expected, the first part of the process, which is strictly ligandbased, is highly reliable and fast. The use of multiple similarity metrics is advantageous since various approaches represent chemical similarity differently, and the consensus-based assessment serves as a good basis for more detailed analysis of hits and their corresponding targets and for inspiring creativity in library design. The platform is open for the incorporation of alternate methods, e.g., shape-based screening or pharmacophore fingerprints. The current target ranking, which is based on the number of hits identified, is not optimal, since it favors large congeneric series. Thus, further modifications to the protocols are ongoing work.
It stands to reason that common challenges of structure-based drug design are especially relevant for an automated procedure, and special attention should be given to it. One of issues concerns the assignment of ligand protonation states, which nevertheless can be reliably estimated by modern pKa predictor software. Correct stereochemistry is more problematic. There are cases where stereochemistry of a PDB ligand is ambiguous; stereochemistry of ChEMBL compounds is not always explicitly defined, and library compounds may exist as racemates or with unknown configuration depending on a synthesis route. The best compromise here is to enumerate relevant stereoisomers and to let the binding pocket decide. Finally, we are faced with incorrect bond orders in a PDB ligand and unknown tautomer forms of ChEMBL or library compounds. Tautomerism is an issue still lacking a sound solution. It is dealt by rule-based generation and docking of sets of tautomers.
Another issue concerns the protein preparation step. As we have shown in this paper, automatic preparation will sometimes detect a wrong binding site or not remove small co-crystallized substances. In the current XGlide implementation, all crystal waters are removed. Therefore, a fraction of automatically created results has to be discarded and manually re-processed. Even if there is still room for improvement, we consider that currently XGlide is one of the best solutions for automatic preparation, protein alignment and docking.
The third step of docking and scoring also has its limitations which are well described in the literature. A consistent binding mode of library compounds is a necessary but not sufficient condition for a compound binding to a target, especially since docking scores are often misleading. These issues together with limitations of previous steps, e.g., complete removal of waters that sometimes provide important contacts to a target, are the main pitfalls of the automatic procedure. Thus, close visual inspection by an expert, who is knowledgeable of a target, will finally allow to judge the relevance of results.
One could argue whether it is worth to use an automatic process with its numerous limitations. In our opinion, the advantages by far outweigh the risks. The process allows an expert to set up a query very easily and to put his time on analysis and re-design of the library. Walking through a full process takes about ten to thirty minutes for a set-up, about 4-8 h for data extraction and similarity search, and about 2 to 3 h for docking per crystal structure if parallelized (depending on the amount of compounds to be docked).
A final word to the expected output
With all known algorithmic and data quality limitations a final library and its assignment to a target will always be “only” an educated guess for a library with significantly enhanced chance to hit a target. Nevertheless, we feel that it is worth the effort.
1 During the step-wise preparation of the library sets only representative subsets were kept via first occurance filters. This resulted in data reduction and therefore the final numbers of DOC-IDs per target were always lower than the numbers in the unfiltered dataset. These results in higher numbers of documents retrieved.
References
- Schamberger J, Grimm M, Steinmeyer A, Hillisch A(2011) Bigger Data, Collaborative Tools and the Future of Predictive Drug Discovery.Drug DiscovToday16: 636-641.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ(2001)Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. AdvDrug Deliv Rev46: 3-26.
- Hann MM (2011) Molecular obesity, potency and other addictions in drug discovery.MedChemComm2: 349-355.
- Lobell M, Hendrix M, Hinzen B, Keldenich J, Meier H, et al.(2006)In silico ADMET traffic lights as a tool for the prioritization of HTS hits.ChemMedChem1: 1229-1236.
- Bohacek RS, McMartin C, Guida WC(1996)The art and practice of structure-based drug design: A molecular modeling perspective. Med Res Rev16: 3-50.
- Lovering F, Bikker J, Humblet C(2009)Escape from Flatland: Increasing Saturation as an Approach to Improving Clinical Success. J Med Chem52: 6752-6756.
- WelschME, SnyderSA, StockwellBR(2010)Privileged scaffolds for library design and drug discovery. CurrOpinChemBiol14: 347-361.
- Khanna K (2012)Drug discovery in pharmaceutical industry: productivity challenges and trends.Drug Discov Today17: 1088-1102.
- Azzaoui K, Hamon J, Faller B, Whitebread S, Jacoby E, et al. (2007)Modeling promiscuity based on in vitro safety pharmacology profiling data. ChemMedChem2: 874-880.
- Huggins DJ, Sherman W, Tidor B(2012)Rational Approaches to Improving Selectivity in Drug Design.J Med Chem55: 1424-1444.
- Ashburn TT, Thor KB(2004) Drug repositioning: identifying and developing new uses for existing drugs.Nat Rev Drug Discov3: 673-683.
- Liu Z, Fang H, Reagan K, Xu X, MendrickDL (2013) In silico drug repositioning: what we need to know. Drug DiscovToday18: 110-115.
- Ekins S, WilliamsAJ, KrasowskiMD, FreundlichJS(2011) In silico repositioning of approved drugs for rare and neglected diseases.Drug DiscovToday16: 298-310.
- RothBL, ShefflerDJ, KroezeWK (2004) Magic shotguns versus magic bullets: selectively non-selective drugs for mood disorders and schizophrenia.Nat Rev Drug Discov3: 353-359.
- Medina-FrancoJL, GiulianottiMA,WelmakerGS, HoughtenRA(2013)Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov Today18: 495-501.
- Bottegoni G, FaviaAD, Recanatini M, Cavalli A(2012)The role of fragment-based and computational methods in polypharmacology. Drug Discov Today17: 23-34.
- Jenwitheesuk E, HorstJA, RivasKL, Van VoorhisWC, Samudrala R(2008)Novel paradigms for drug discovery: computational multitarget screening. Trends PharmacolSci29: 62-71.
- SchomburgKT, Bietz S, Briem H, HenzlerAM, Urbaczek S, et al. (2014)Facing the challenges of structure-based target prediction by inverse virtual screening. J Chem Inf Model54: 1676-1686.
- PetronePM, Simms B, Nigsch F, Lounkine E, Kutchukian P, et al.(2012)Rethinking molecular similarity: comparing compounds on the basis of biological activity. ACS Chem Biol7: 1399-1409.
- Riniker S, Wang Y, JenkinsJL, LandrumGA(2014)Using information from historical high-throughput screens to predict active compounds.J Chem InfModel54: 1880-1891.
- ChEMBL (2016) Available from: https://www.ebi.ac.uk/chembl/ (Accessed on: January 05, 2018).
- Protein Data Bank (2016)A Structural View of Biology.Available from: https://www.rcsb.org/pdb/home/home.do (Accessed on: January 05, 2018).
- Johnson M, Maggiora GM (1990)Concepts and Applications of Molecular Similarity. John Wiley and Sons, New York, USA.
- Pipeline Pilot, Accelrys Software Inc. (2013)BIOVIA Pipeline Pilot.
- GaultonG, BellisLJ, BentoAP, Chambers J, Davies M, et al. (2012) ChEMBL: a large-scale bioactivity database for drug discovery.Nucleic Acids Res40: D1100-D1107.
- BaberJC, ShirleyWA, Gao Y, Feher M (2006)The Use of Consensus Scoring in Ligand-Based Virtual Screening.J ChemInf Model46: 277-288.
- Whittle M, Gillet VJ, Willett P, Loesel J(2006)Analysis of Data Fusion Methods in Virtual Screening: Theoretical Model. J ChemInf Model46: 2193-2205.
- Rogers Hahn M (2010) Extended-connectivity fingerprints.J Chem Inf Model50: 742-754.
- CramerRD, JilekRJ, AndrewsKM (2002)Topomer similarity searching of conventional structure databases.J Mol Graph Model20: 447-462.
- Rarey M, DixonJS (1998) Feature trees: A new molecular similarity measure based on tree matching. J ComputAided Mol Des12: 471-490.
- Fraczkiewicz R, Lobell M, GöllerAH, Krenz U, Schoenneis R, et al. (2015) Best of both worlds: combining pharma data and state of the art modeling technology to improve in Silico pKa prediction. J Chem Inf Model55: 389-397.
- ADMET Predictor(2014) Simulations Plus, Lancaster, CA, 93534, USA.
- Flipo M, Desroses M, Lecat-Guillet N, Villemagne B, Blondiaux N, et al.(2012)Ethionamide boosters. 2. Combining bioisosteric replacement and structure-based drug design to solve pharmacokinetic issues in a series of potent 1,2,4-oxadiazole EthR inhibitors. J Med Chem55: 68-83.
- UniProt (2016)Available from: https://www.uniprot.org (Accessed on: January 05, 2018).
- PDB Bind (2014)Available from: https://www.pdbbind-cn.org (Accessed on: January 05, 2018).
- HalgrenTA(2009)Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model49: 377-389.
- Doré S, Okrasa K, PatelJC, Serrano-Vega M, Bennett K, et al.(2014)Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature511: 557-562.
- Wang WH, Katritch V, HanGW, HuangXP, Liu W (2013) Structure of the human smoothened receptor bound to an antitumour agent.Nature497: 338-343.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences