Three dimensional structure modeling of lanosterol 14-alpha demethylase of Candida albicans and docking studies with new triazole derivatives
Mohammad Irfan, Mohammad Abid
Mohammad Irfan, Mohammad Abid*
Medicinal Chemistry Laboratory, Department of Biosciences, Jamia Millia Islamia (A Central University), New Delhi-110025, India
- *Corresponding Author:
- Mohammad Abid
Medicinal Chemistry Laboratory
Department of Biosciences, Jamia Millia Islamia (A Central University)
New Delhi-110025
India
Tel: +91-8750295095
Fax: +91-11-2698 0229
E-mail:mabid@jmi.ac.in
Received date: June 29, 2015; Accepted date: August 28, 2015; Published date: August 31, 2015
Citation: Irfan M, Abid M. Three dimensional structure modeling of lanosterol 14-α demethylase of Candida albicans and docking studies with new triazole derivatives. Chem Inform. 2015, 1:1.
Abstract
Objective: In this report, three dimensional structure of lanosterol 14-α demethylase (CYP51) protein of Candida albicans was modeled and used for docking studies with eighteen synthetic triazole derivatives. Methods: The model was generated by multiple threading alignment and iterative structure assembly simulation by I-TASSER and validated by Ramachandran plot using PROCHECK and MolProbity servers. The eighteen 1,2,3-triazole compounds were previously synthesized using two different synthetic routes by our research group in the search of new antifungal agents. The binding affinity and interaction of all triazole compounds with predicted model of CYP51 were determined by AutoDock Vina. Results: I-TASSER generated five models in which Model 1 was selected as best predicted structure on the basis of C-score 1.30, TM-score 0.89 ± 0.07, and RMSD 4.7 ± 3.1 Å. It showed 89.2% residues in most favoured region and 7.1% residues in additional allowed regions. Docking results showed that all the derivatives have good binding energy in the range -9.8 to -7.4 kcal/mol. Moreover, compound NT- 03 (-9.8 kcal/mol) was found as best inhibitor as it binds nicely into the active site of the enzyme. Conclusion: The study suggests that I-TASSER is very easy and reliable tool for three dimensional structure predictions. The N-atom in quinoline ring of compound NT- 03 involved in the additional interaction with predicted model of CYP51. Finally, I-TASSER might be used as an important tool in drug designing process in search of more effective therapies.
https://bluecruiseturkey.co
https://bestbluecruises.com
https://marmarisboatcharter.com
https://bodrumboatcharter.com
https://fethiyeboatcharter.com
https://gocekboatcharter.com
https://ssplusyachting.com
Keywords
Lanosterol 14-α demethylase; homology modelling; I-TASSER; Docking studies; Triazoles; Candida albicans
Introduction
The increased incidences of candidiasis and alarming rate of drug resistance has prompted researchers to develop novel and more effective antifungal agents. Opportunistic yeast Candida, mainly C. albicans, is responsible for this complicated fungal infection with 50-90% human candidiasis [1,2]. Commonly used antifungal drugs belong to various classes and characterised by different mechanisms of action and spectrum of activity. Among them, azoles are the first choice of drugs for the treatment of fungal infection [3]. They interfere with the synthesis of ergosterol by inhibiting CYP51 enzyme of its biosynthetic pathway in C. albicans [4]. CYP51, a member of Cytochrome P450 family, is a heme thiolate enzyme which converts lanosterol into 4,4'-dimethyl cholesta-8,14,24-triene-3-beta-ol [5]. The activity of azole drugs is attributed to the co-ordination binding of the heterocyclic nitrogen atom (N-3 of imidazole and N-4 of triazole) to the heme iron atom in the binding site of CYP51 enzyme. Due to inhibition of CYP51, depletion of ergosterol coupled with an accumulation of 14-methyl sterols which results in impaired fungal cell growth [6].
make it an important target for antifungal drug design [7]. Although the advance in bioinformatics tools may help to develop new drug molecules, the absence of crystal structure of CYP51 enzyme of C. albicans is a major obstacle in the drug design efforts. Currently 38 X-ray coordinates of CYP51 available in PDB in which, one is for Saccharomyces cerevisiae (4LXJ) [8] and others for non-membrane bound CYP51 like Trypanosoma brucei (3G1Q) [9], Mycobacterium tuberculosis (2BZ9) [10], Homo sapiens (3JUV) [11], Leishmania infantum (3L4D) [12] etc. In recent years, the advance research in computer algorithms for three dimensional structure and function prediction have resolved this problem to an extent and helps to obtain valuable information of desired protein [13]. The three dimensional structure prediction by computational methods has divided into three categories: i) comparative modeling based on evolutionarily related homologous template [14], ii) threading methods which use template of proteins from different evolutionary origin with similar sequences [15], and iii) ab initio modeling in which no structural similarities for query protein available in PDB [16,17].
In this study, we selected I-TASSER (iterative threading assembly refinement) server for 3D structure prediction of CYP51 enzyme of C. albicans. I-TASSER has composite approaches of threading, structural refinement, and ab initio modeling to generate reliable model which is different from earlier reported methods of CYP51 structure prediction [18]. I-TASSER has been ranked as the best server for automated protein structure prediction in the last five consecutive CASP7 [19], CASP8 [20], CASP9 [21], CASP10 [22], and CASP11 experiments. The predicted model was used for the purpose of docking studies with eighteen newly synthesized triazoles derived from naturally bioactive scaffolds to explore their interactions and binding affinity in search of more potent drug like molecules.
Methods
Three dimensional structure prediction by I-TASSER
The sequence of 528 amino acids of C. albicans lanosterol 14-α demethylase protein (CYP51) was retrieved form Swiss Prot database (https://www.uniprot.org/uniprot/P10613) in FASTA format as follows:
MAIVETVIDGINYFLSLSVTQQISILLGVPFVYNLVWQYLYSLRKDRAPLVFYWIPWFGS AASYGQQPYEFFESCRQKYGDVFSFMLLGKIMTVYLGPKGHEFVFNAKLSDVSAEDAYKHLTTPVFGKGVIYDCPNSRLMEQKKFAKFALTTDSFKRYVPKIREEILNYFVTDESFKLKEKTHGV ANVMKTQPEITIFTASRSLFGDEMRRIFDRSFAQLYSDLDKGFTPINFVFPNLPLPHYWRRDAAQKKISATYMKEIKSRRERGDIDPNRDLIDSLLIHSTYKDGVKMTDQEIANLLIGILMGGQ HTSASTSAWFLLHLGEKPHLQDVIYQEVVELLKEKGGDLNDLTYEDLQKLPSVNNTIKETLRMHMPLHSIFRKVTNPLRIPETNYIVPKGHYVLVSPGYAHTSERYFDNPEDFDPTRWDTAAAK ANSVSFNSSDEVDYGFGKVSKGVSSPYLPFGGGRHRCIGEQFAYVQLGTILTTFVYNLRWTIDGYKVPDPDYSSMVVLPTEPAEIIWEKRETCMFGTILTTFVYNLRWTIDGYKVPDPDYSSMVV LPTEPAEIIWEKRETCMF
The three dimensional model was generated using I-TASSER server which generates 3D model of query sequence by multiple threading alignments and iterative structural assembly simulation [17]. The reasons behind the selection of this server were its availability, composite approach of modeling and performance in CASP competition. I-TASSER methodology includes general steps of threading, structural assembly, model selection, refinement, and structure based functional annotation [18]. Firstly, the evolutionary related protein against query sequence was checked by running PSIBLAST which created sequence profile and used to predict secondary structure by PSIPRED [23]. Then, the query sequence was threaded through the representative PDB structure library using LOMETS [24]. The quality of template alignment was checked by Z-score and the best one used for the further consideration. In next step, the continuous fragments in threading alignments were excised to from an assemble structure model of aligned regions. The structural assembly of unaligned regions was built by ab initio modeling [17]. The modeling accuracy of unaligned regions is generally low while threading aligned regions have high accuracy so these template fragments keep rigid in simulation process to obtained high resolution structure. The replica exchange Monte Carlo simulation technique was used for fragment assembly [25]. The simulation include Cα/side chain correlation, H-bonds, hydrophobicity, spatial restrains from threading templates [24], and sequence based contact predictions from SVMSEQ [26]. The conformations generated during refinement simulation process were clustered by SPICKER [27] and the average of three dimensional coordinates of all the clustered structure was calculated to obtained cluster centroids.
In refinement process, the selected cluster centroids were again used to perform further fragment assembly simulation which helps to remove steric clashes and to refine the global topology of the cluster centroids. The PDB structures, structurally closed to the cluster centroids, were identified by TM-align [28]. The final structural models were generated by REMO [29] in which cluster centroids of second round simulation used as input. In last step, the function of three dimensional model of query protein was predicted by matching the proteins of known structure and function in PDB. The functional analogs were ranked on the basis of TM-score, RMSD, sequence identity, and coverage of the structure alignment. The quality of predicted model was determined by C-score (confidence score) which is ranged as −5 to 2. It depends on quality of threading alignment and the convergence of structural assembly refinement simulations.
Validation of predicted 3D model (model 1)
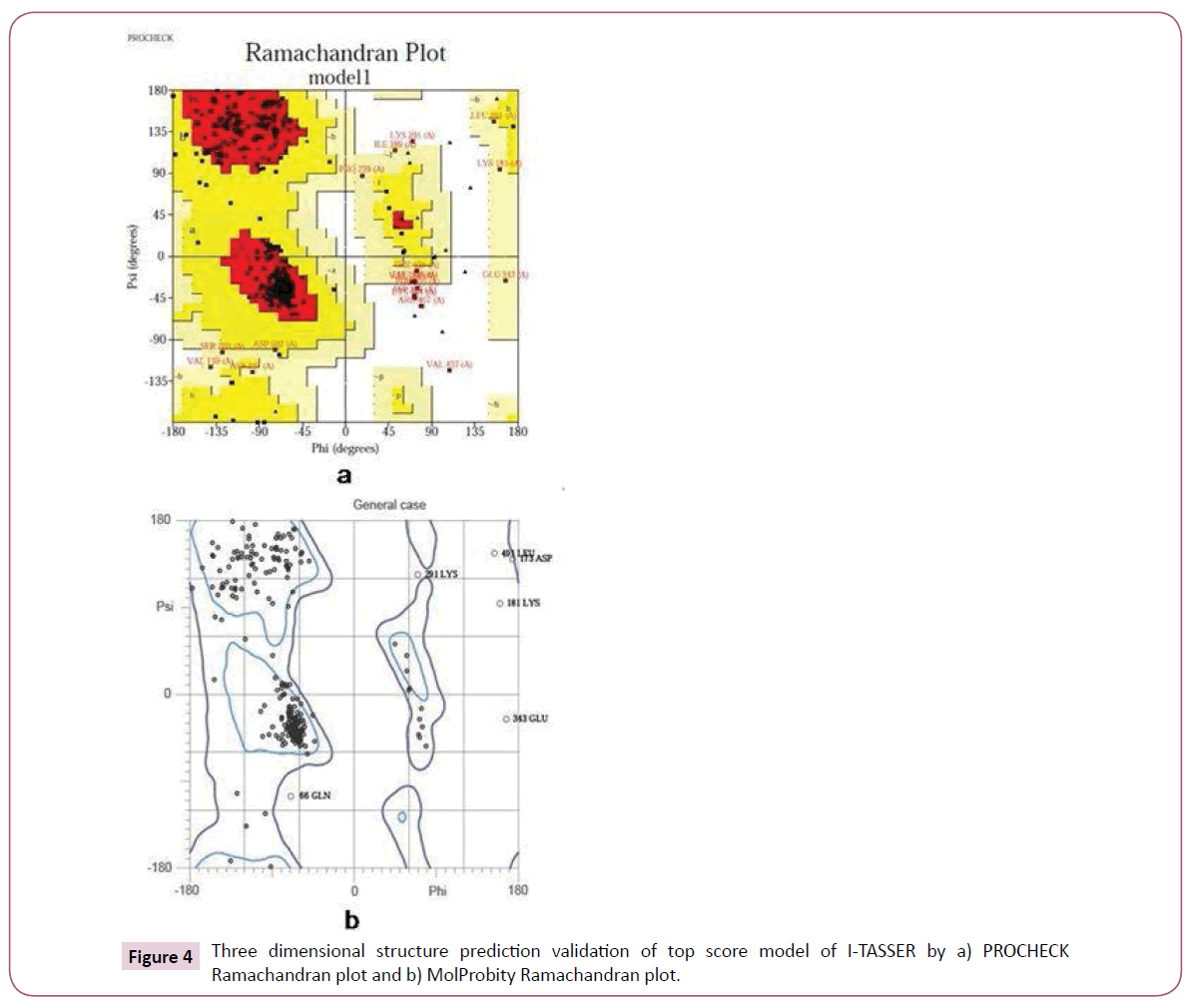
The conformation of best 3D model (model 1) predicted by I-TASSER was further validated by Ramachandran plot. The conformation of the predicted model was calculated by analyzing the phi (Φ) and psi (Ψ) torsion angles using PROCHECK online server. The Ramachandran plot obtained from PROCHECK describes a good quality model which has over 90% residues in most favoured region. The assessment of model was also confirmed by MolProbity online server [30].
Inhibitors for docking studies
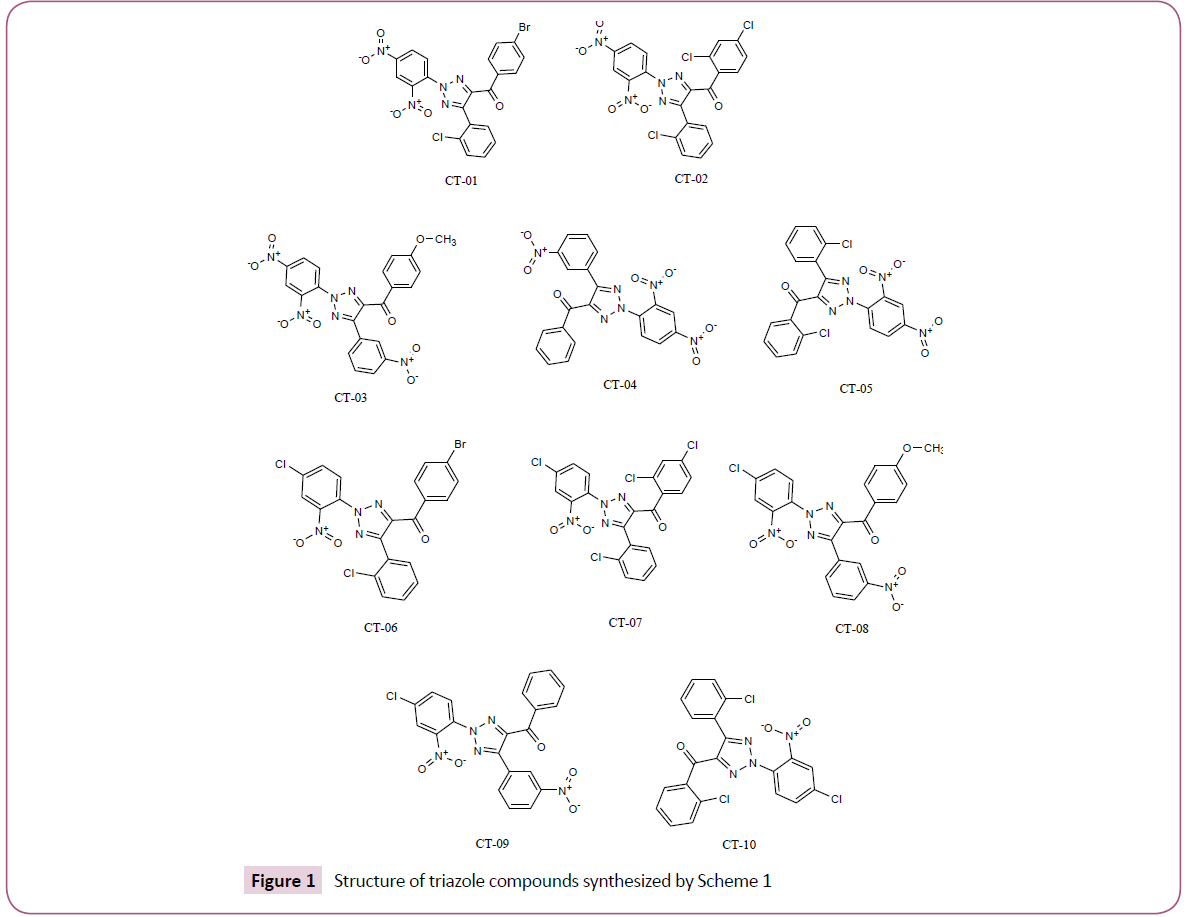
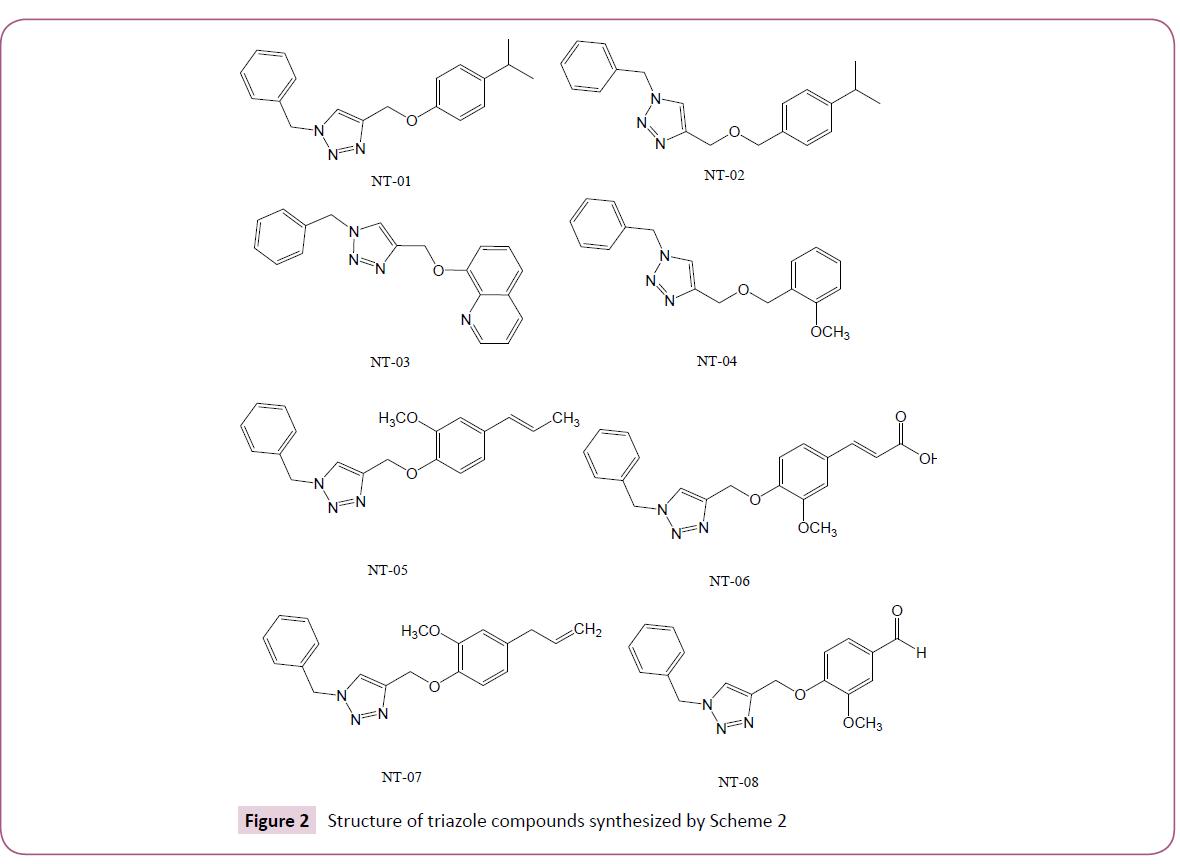
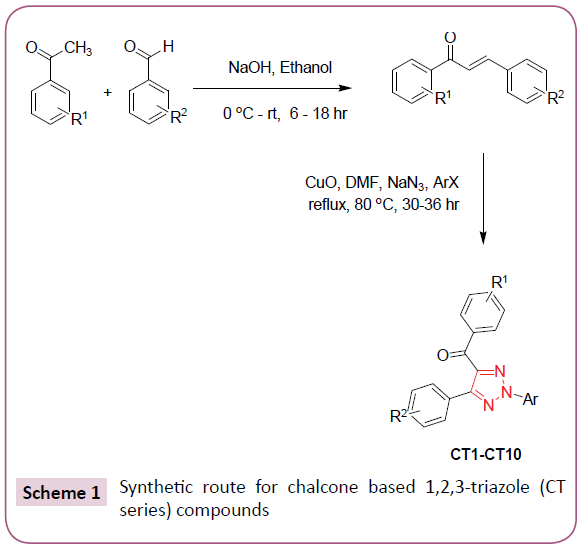
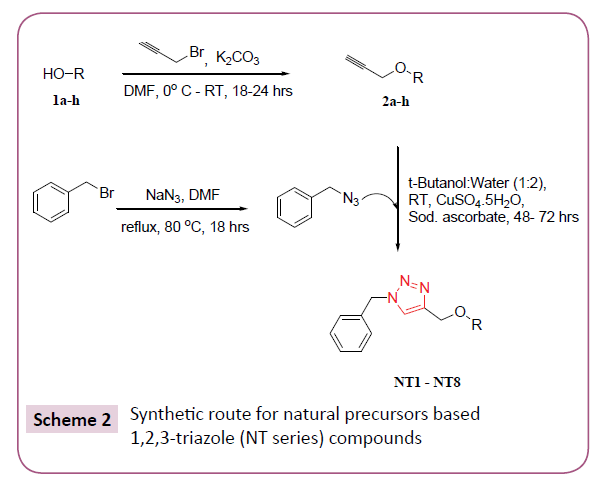
In silico screening of selected triazole derivatives with the predicted model of C. albicans’s CYP51 enzyme was done in order to find their interactions and binding energy. Triazole derivatives used in the study were recently synthesized by our group (Figures 1 and 2). 1,2,3-Triazole functionality showed various biological activities and present in different antifungal drugs like fluconazole, itraconazole, voriconazole etc [31]. Compounds CT-01 to CT-10 were synthesised by one pot three-component stepwise approach based on azide-chalcone oxidative cycloaddition and post-triazole arylation (Scheme 1) [32]. While compounds NT-01 to NT-08 was obtained by modification of some naturally occurring alcohols using [32] azide-alkyne cycloaddition strategy (Scheme 2) [33].
Docking studies
In this study, AutoDock Vina was used to determine the binding energy of designed triazole derivatives with predicted model obtained from I-TASSER and compared with the standard drug, fluconazole. AutoDock Vina is advanced version which is much faster and accurate in blinding mode prediction if compared with its older version AutoDock 4.0 [34]. The affinity grid box was centred to the binding site and the grid spacing fixed to 0.375 Å which is highly recommended. The program automatically calculates the grid map and grid centre coordinates (X=-2.0, Y=0.98, Z=0.41) considering the predicted binding site of enzyme. After generating receptor and all ligands file in PyMOL AutoDock/ Vina plugin, Vina was ran to obtained docking score (binding affinity in kcal/mol) and RMSD value for all compounds. PyMOL was used for visualization of the polar contacts in between ligands and binding site of protein [35]. The best docking result can be considered to be the conformation with the lowest docking score or binding energy and lowest RMSD.
Results and Discussion
Three dimensional structure prediction
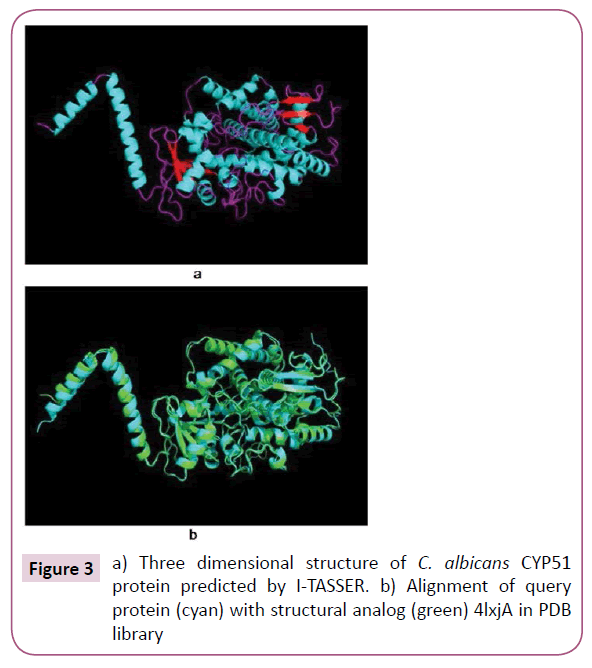
The predicted model of C. albicans CYP51 protein and its three dimensional coordinate file in PDB format were successfully obtained from I-TASSER. The results obtained from server includes predicted secondary structure with confidence score (range 0 to 9), predicted solvent accessibility, five predicted structures with C-score, top ten template from PDB used in alignment, top ten PDB structural analogs, functional analogs protein, and binding site residues. Model 1 (Figure 3) was selected as best predicted model with C-score 1.30, TM-score 0.89 ± 0.07, and RMSD 4.7 ± 3.1 Å. C-score with higher value reflect a model of better quality [18].
Top ten threading templates for query protein sequence were identified by LOMETS meta-server (Table 1). The threading alignment is generally estimated by normalized Z-score. However, a normalized Z-score >1 value reflect a confident alignment but in case of small alignment of large query sequence, it does not give significant indication of modeling accuracy. Then percentage sequence identity in the threading aligned region (Iden1) and in the whole chain (Iden2) considered for the good homology (Table 1). The structural alignment program, TM-align, identified 4lxjA in PDB library as best structural analog of the top scoring model of I-TASSER with the TM-score of 0.975 (Table 2). The ligand binding site of Model 1 was predicted and aligned with the template (best hit 3juvA) binding site by TM-align. The binding score (BS) and confidence score of ligand binding (CscoreLB) for predicted binding site with template (3juvA) were measured as 1.38 and 0.73. The BS score is based on structure similarity between binding site of template and predicted model of I-TASSER. The BS score >1 and CscoreLB >0.5 indicates more reliable ligand binding site prediction [18].
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Z-score |
|---|---|---|---|---|---|
| 1 | 4k0fA | 0.65 | 0.64 | 0.98 | 4.09 |
| 2 | 4k0fA | 0.65 | 0.64 | 0.98 | 4.45 |
| 3 | 1e9xA | 0.27 | 0.25 | 0.84 | 4.11 |
| 4 | 4k0fA | 0.64 | 0.64 | 0.98 | 5.06 |
| 5 | 1e9xA | 0.27 | 0.25 | 0.85 | 3.31 |
| 6 | 4k0fA | 0.64 | 0.64 | 0.98 | 3.74 |
| 7 | 4k0fA | 0.64 | 0.64 | 0.98 | 3.68 |
| 8 | 4lxjA | 0.64 | 0.64 | 0.98 | 1.85 |
| 9 | 4k0fA | 0.64 | 0.64 | 0.98 | 5.98 |
| 10 | 4k0fA | 0.65 | 0.64 | 0.98 | 6.72 |
Table 1: Top ten templates used by I-TASSER for threading alignment. Iden1 is the percentage sequence identity of the template used by I-TASSER. Iden2 is the percentage sequence identity of the whole template chain with query sequence. Cov. represents the coverage of the threading alignment and is equal to the number of aligned residues divided by the length of query protein.
| Rank | PDB Hit | TM-score | RMSDa | IDEN | Cov. |
|---|---|---|---|---|---|
| 1 | 4lxjA | 0.975 | 0.85 | 0.642 | 0.985 |
| 2 | 3jusB | 0.805 | 2.02 | 0.396 | 0.841 |
| 3 | 2x2nD | 0.775 | 2.31 | 0.295 | 0.826 |
| 4 | 2wuzB | 0.772 | 2.22 | 0.313 | 0.822 |
| 5 | 1tqnA | 0.762 | 3.27 | 0.177 | 0.854 |
| 6 | 1bu7B | 0.751 | 3.04 | 0.145 | 0.835 |
| 7 | 2ve3A | 0.745 | 2.86 | 0.196 | 0.818 |
| 8 | 1n6bA | 0.741 | 3.20 | 0.168 | 0.835 |
| 9 | 2vn0A | 0.740 | 3.27 | 0.161 | 0.837 |
| 10 | 2hi4A | 0.736 | 3.58 | 0.175 | 0.849 |
Table 2: Top ten structural analogs in PDB identified by TM-align. RMSDa is the RMSD between residues that are structurally aligned by TM-align. IDEN is the percentage sequence identity in the structurally aligned region. Cov. represents the coverage of the alignment by TM-align and is equal to the number of structurally aligned residues divided by length of the query protein.
| Code | Binding energy (kcal/mol) | RMSD | Code | Binding energy (kcal/mol) | RMSD |
|---|---|---|---|---|---|
| CT-01 | −8.4 | 0 | CT-10 | −8.4 | 0 |
| CT-02 | −9.1 | 0 | NT-01 | −9.1 | 0 |
| CT-03 | −8.7 | 0 | NT-02 | −9.7 | 0 |
| CT-04 | −8.4 | 0 | NT-03 | −9.8 | 0 |
| CT-05 | −8.7 | 0 | NT-04 | −8.7 | 0 |
| CT-06 | −7.4 | 0 | NT-05 | −9.4 | 0 |
| CT-07 | −8.6 | 0 | NT-06 | −8.9 | 0 |
| CT-08 | −8.3 | 0 | NT-07 | −9.1 | 0 |
| CT-09 | −8.9 | 0 | NT-08 | −9.3 | 0 |
| FLC | −8.6 | 0 |
Table 3: Binding affinity of docked compounds with predicted model of Candida albicans lanosterol 14-α demethylase protein. FLC=Fluconazole.
Assessment of predicted model
The Ramachandran plots of best predicted model were obtained from PROCHECK and MolProbity servers which showed the reliability of model. The PROCHECK Ramachandran plot showed 89.2% residues in most favoured regions and 7.1% residues in additional allowed regions i.e., the total of 96.3% residues in allowed regions which indicates a good quality model. MolProbity Ramachandran plot also showed 96.6% residues in allowed regions which again confirmed the quality of predicted model (Figure 4).
Docking studies
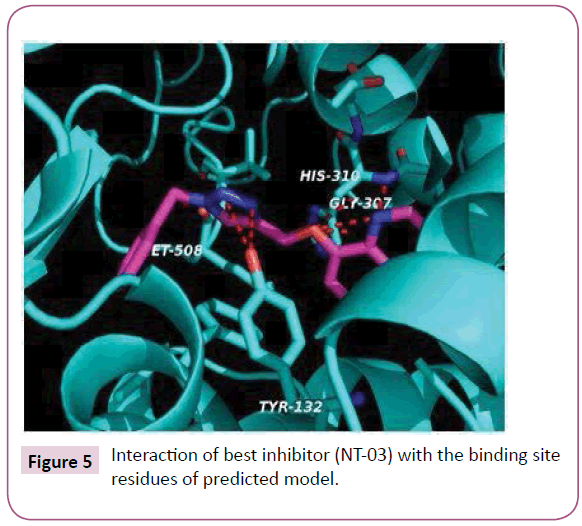
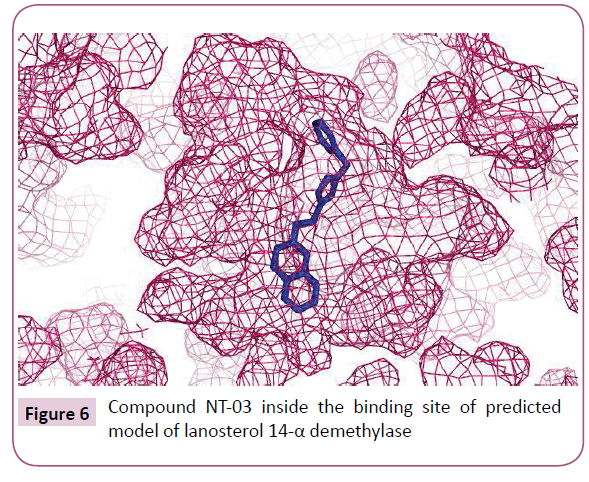
The molecular docking of all the triazole derivatives with best predicted model of C. albicans CYP51 protein was successfully completed using AutoDock Vina. All the docked compounds showed good binding energy with a range of -9.8 to -7.4 kcal/mol best compound in its series with lowest binding energy -9.1 kcal/ mol. However all the compounds of scheme 2 showed less binding energy than the fluconazole and compound NT-03 with binding energy -9.8 kcal/mol was selected as best inhibitor. It also showed good interactions with the binding site residues of target protein. The N-atom of triazole ring was interacted with the TYR-132 residue of binding site. Compound NT-03 having quinoline ring also showed additional interactions with GLY-307 and HIS-310 residues in binding pocket due to N-atom of the quinoline ring (Figures 5 and 6).
Conclusion
In summary, three dimensional model of C. albicans CYP51 protein was built by multiple threading alignment and iterative structure assembly simulation by I-TASSER. The crystal coordinate 4lxjA in PDB library was found very close structural resemblance with predicted model. The reliability and accuracy of model were confirmed by Ramachandran plot using PROCHECK and MolProbity server. The docking studies with predicted modelshowed that compound NT-03 has good binding affinity (-9.8 kcal/mol). Moreover, it interacted with TYR-132 and GLY-307 and HIS-310 residues in binding site of predicted protein which is in accordance with the experimental bioactivity data. It can be concluded that natural compound based triazole derivatives can be developed as more effective antifungal agent.
Acknowledgement
Mohammad Abid gratefully acknowledges the funding support in the form of Major Research Project by University Grant Commission (UGC), Govt. of India (Grant No. 41-277/2012 (SR)). MI would like to acknowledge UGC, INDIA for fellowship.
Conflict of interest
Authors declare no conflict of interest.
References
- Vázquez?González D, Perusquía?Ortiz AM, Hundeiker M, Bonifaz A (2013) Opportunistic yeast infections: candidiasis, cryptococcosis, trichosporonosis and geotrichosis. J DtschDermatolGes11: 381-394.
- Wächtler B, Citiulo F, Jablonowski N, Förster S, Dalle F, et al. (2012) Candida albicans-epithelial interactions: dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PloS one 7: e36952.
- Mansfield BE, Oltean HN, Oliver BG, Hoot SJ, Leyde SE, et al. (2010) Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoSPathog 6: e1001126.
- Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, et al. (2009) Efflux-mediated antifungal drug resistance. ClinMicrobiol Rev22: 291-321.
- Bard M, Lees ND, Turi T, Craft D, Cofrin L, et al. (1993) Sterol synthesis and viability oferg11 (cytochrome P450 lanosteroldemethylase) mutations in Saccharomyces cerevisiae and Candida albicans.Lipids 28(11): 963-967.
- Ji H, Zhang W, Zhang M, Kudo M, Aoyama Y, et al. (2003) Structure-Based de Novo Design, Synthesis, and Biological Evaluation of Non-Azole Inhibitors Specific for Lanosterol 14α-Demethylase of Fungi. J Med Chem 46: 474-485.
- Lamb DC, Kelly DE, Venkateswarlu K, Manning NJ, Bligh HF, et al. (1999) Generation of a Complete, Soluble, and Catalytically Active Sterol 14α-Demethylase-Reductase Complex. Biochemistry 38: 8733-8738.
- Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JD, et al. (2014) Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. ProcNatlAcadSci111: 3865-3870.
- Lepesheva GI, Park HW, Hargrove TY, Vanhollebeke B, Wawrzak Z, et al. (2010) Crystal structures of Trypanosomabrucei sterol 14α-demethylase and implications for selective treatment of human infections. J BiolChem285(3): 1773-1780.
- Podust LM, von Kries JP, Eddine AN, Kim Y, Yermalitskaya LV, et al. (2007) Small-molecule scaffolds for CYP51 inhibitors identified by high-throughput screening and defined by X-ray crystallography.Antimicrob Agents Chemother 51: 3915-3923.
- Strushkevich N, Usanov SA, Park HW (2010) Structural basis of human CYP51 inhibition by antifungal azoles. J MolBiol397: 1067-1078.
- Hargrove TY, Wawrzak Z, Liu J, Nes WD, Waterman MR, et al. (2011) Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14α-demethylase (CYP51) from Leishmaniainfantum. J BiolChem 286: 26838-26848.
- Zhang Y (2008) Progress and challenges in protein structure prediction. CurrOpinStructBiol 18: 342-348.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res25: 3389-3402.
- Jones DT, Taylort WR, Thornton JM (1992) A new approach to protein fold recognition. Nature 358: 86-89.
- Simons KT, Kooperberg C, Huang E and Baker D (1997) Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J MolBiol 2689: 209-225.
- Wu S, Skolnick J, Zhang Y (2007) Ab initio modeling of small proteins by iterative TASSER simulations. BMC biol5: 17.
- Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc5: 725-738.
- Battey JND, Kopp J, Bordoli L, Read RJ, Clarke ND et al. (2007) Automated server predictions in CASP7. Proteins69: 68-82.
- Zhang Y (2009) I-TASSER: Fully automated protein structure prediction in CASP8. Proteins 77: 100-113.
- Moult J, Fidelis K, Kryshtafovych A, Tramontano A (2011) Critical assessment of methods of protein structure prediction (CASP)-round IX. Proteins 79: 1-5.
- Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A (2014) Critical assessment of methods of protein structure prediction (CASP)-round X. Proteins: Struct, Funct, Bioinf 82: 1-6.
- Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J MolBiol 292: 195-202.
- Wu S, Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35: 3375-3382.
- Zhang Y, Kihara D, Skolnick J (2002) Local energy landscape flattening: parallel hyperbolic Monte Carlo sampling of protein folding. Proteins 48: 192-201.
- Wu S, Zhang Y (2008) A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics24: 924-931.
- Zhang Y, Skolnick J (2004) SPICKER: A clustering approach to identify near-native protein folds. J ComputChem 25: 865-871.
- Zhang Y, Skolnick J (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res 33: 2302-2309.
- Li Y, Zhang Y (2009) REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins 76: 665-676.
- Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, et al. (2003) Structure validation by Cα geometry: Φ, Ψ and Cβ deviation. Proteins50: 437-450.
- Khan K M, Siddiqui S, Saleem M, Taha M, Saad SM, et al. (2014) Synthesis of triazole Schiff bases: Novel inhibitors of nucleotide pyrophosphatase/phosphodiesterase-1. Bioorg Med Chem 22: 6509-6514.
- Irfan M, Khan M, Manzoor N, Abid M (2014) Synthesis of N-2-aryl-substituted-1, 2, 3-triazole Derivatives as Novel Inhibitors of Entamoebahistolytica. OJOC2: 21-28.
- Irfan M, Aneja B, Yadava U, Khan SI, Manzoor N, et al. (2015) Synthesis, QSAR and anticandidal evaluation of 1, 2, 3-triazoles derived from naturally bioactive scaffolds.Eur J Med Chem93: 246-254.
- Trott O, Olson AJ (2010) AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J ComputChem 31: 455-461.
- DeLano WL (2002) ThePyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, USA, 2002.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences