Ligand Based Pharmacophore Modeling, Virtual Screening and Molecular Docking for Identification of Novel CYP51 Inhibitors
Sarvesh Kumar P, Aarti Singh, Swapnil Sharma, Mukta Sharma and Anupama Mittal
Sarvesh Kumar P, Aarti Singh*, Swapnil Sharma, Mukta Sharma and Anupama Mittal
Department of Pharmacy, Banasthali University, Rajasthan, India
- *Corresponding Author:
- Aarti Singh
Department of Pharmacy, Banasthali University
PO Banasthali
Rajasthan-304 022
India
E-mail: modernchem.aarti@gmail.com
Received date: September 09, 2015; Accepted date: October 01, 2015; Published date: October 08, 2015
Citation: Sarvesh Kumar P, Singh A, Sharma S. Ligand Based Pharmacophore Modeling, Virtual Screening and Molecular Docking for Identification of Novel CYP51 Inhibitors. Chem Inform. 2015, 1:2.
Abstract
Background: Azoles were designed to impede fungal infection by inhibiting the prominent target Lanosterol 14α-demethylase (CYP51), an essential component of fungal cell membrane, in turn interrupting the pathway of lanosterol to ergosterol. Emerging resistance ability and toxicity of presently available drugs urge for novel antifungal agents. Methods and findings: This research focus on cost effective ligand based pharmacophore modeling to present potential novel CYP51 inhibitors. Out of 10 hypotheses, the best 4 featured pharmacophore model- 3HBA and 1HY with RMS 0.85, correlation coefficient of 0.94 and cost difference of 33.33 has been chosen. The prediction of generated pharmacophore was validated using rm2 matrices, Fischer’s randomization method, internal and external test prediction. The chosen model was used as a 3D query to screen the chemical databases (NCI and Maybridge). Two hits (NSC 1028 and HTS 00684) with high fit value and zero Lipinski’s violation were docked into CYP51 receptor site to identify key amino acid residues. Conclusions: Two hits (NSC 1028 and HTS 00684) with high fit value (7.56 and 8.25) and low estimated value (0.16 and 0.03) were docked into the catalytic site of CYP51 receptor. Hydrogen bond, Van Der Waals and hydrophobic interactions governed the binding mode of the hits within the active site. The results showed high affinity of hits when compared with the standard drug (fluconazole and voriconazole) which directs future evaluation of these hits using in vivo studies.
Keywords
Azoles; Ligand based pharmacophore modeling; Virtual screening; Molecular docking
Introduction
Candida albicansis an opportunistic pathogen that resides in the gastrointestinal and genitourinary tracts in about 70% of humans in commensalism. In immunocompromised patients and some immunologically weak individuals, it becomes opportunistic pathogen leading to the infection candidiasis [1,2]. An assortment of non-albicans species like Candida parapsilosis, Candida tropicalis, Candida krusei and Candida glabrata are recovered from infected patients nevertheless Candida albicans remains a major infectious fungal agent. There is a dramatic elevation in invasive fungal infections and associated mortality rate. Candida albicans has emerged as an influential member of model organisms mainly for fungal pathogens.
Introduction of azole antifungal agents such as amphotericin B, fluconazole and voriconazole had greatly facilitated the treatment of C. albicans infections. As these antifungal agents are soluble in water and have high degree of bioavailability after oral administration so has been used extensively to treat wide range. The azoles are targeted primarily on inhibition of ergosterol synthesis through an interaction with lanosterol 14-alpha demethylase (CYP51), an enzyme dependent on cytochrome P-450 that is necessary for the conversion of lanosterol to ergosterol [3]. The depletion of ergosterol directed the increase in cell membrane permeability and inhibition of cell growth and replication.
Cytochrome P450 isoenzyme 51 (CYP51/lanosterol 14α-demethylase) is a family of phylogenetic well preserved monooxygenases found in mycobacteria, fungi, plants, animals and humans. CYP51 catalyses the oxidative removal of the 14α-methyl group (C-32) of lanosterol to give desaturated intermediates during ergosterol biosynthesis. Three successive monooxygenation reactions within this catalytic cycle results in the formation of 14-hydroxy-methyl, 14-carboxaldehyde, and 14-formyl derivatives further followed by elimination of formic acid with simultaneous introduction of C14, C15 double bond [4-6]. Antifungal agents ought to be potent inhibitors of fungal CYP51 leaving human CYP51 unaffected. In extensive structureactivity studies, the azole ring has been considered as one of the most vital pharmacophores for antifungal activity. Azoles are considered because of their in vivo efficacy, their broader spectrum of activity, better bioavailability and their apparent fungicidal activity in some infections [7,8]. Concomitant with their widespread use, there has been conspicuous elevation in the rate of toxicity and antifungal resistance resulting in urgent need for authentically broad spectrum and low-toxic antifungal agents [9].
Our approach deals with the development of an accurate and efficient method for discovering putative CYP51 inhibitors. The pharmacophore hypothesis generation was defined by prominent chemical features of compounds with CYP51 inhibitory activity. The generated hypothesis offers a rational hypothetical view of these important chemical features responsible for activity. On the basis of structure-activity relationship, a ligand-based 3Dimensional pharmacophore hypothesis was generated [10]. Various validation methods have been employed on the generated hypothesis such as rm2 metrics, Fischer’s randomization method, internal test set method, and external test set method. This validated hypothesis was used for high throughput screening to sort out hits. The retrieved hits were subjected to varied refining procedures based on fit value, estimated value, and Lipinski’s rule of five. Molecular docking has been performed to further explain the drug-likeness prediction of retrieved hits.
Experimental Methods
Dataset collection
The pharmacophore generation demands for an accurate and precise selection of training set. In course of data set selection, following criteria ought to be considered. (1) Binding interactions of the compounds with the receptor characterize their activity; (2) Diversified activity range of at least 4 orders of magnitude; (3) the most and least active compound must be incorporated in the training set and homogenous procedures are carried out to attain all biologically relevant data.
Considering abovementioned criteria, 28 CYP51 inhibitors have been collected to form a database [11]. These compounds were sketched using ChemDraw program version 8.0 which were consequently converted into 3-Dimensional using Discovery Studio 2.0. Afterwards, a vigilant selection of 21 training set compounds and 7 test set compounds has been accomplished.
Diverse conformation generation
Diverse conformation generation protocol executed in Discovery Studio (DS) has been formulated to generate assorted conformations using best conformation model generation. The parameters such as maximum number of 255 conformations, the best conformational analysis and an energy threshold of 20 kcal/mol above the global energy minimum were considered for generating varied conformers [12]. Each compound attains its energy minimized conformation by facilitating CHARMM force field.
Pharmacophore modeling
Pharmacophore hypothesis generation has been conceded with 21 training set compounds. Prior to hypothesis generation, chemical features common amongst active training set compounds were selected using feature mapping protocol implemented in Discovery Studio (DS). The pharmacophore hypotheses were generated by Hypogen module implemented in Discovery Studio (DS) version 2.0. The generated hypotheses were ranked considering their best correlation with the 3-Dimensional spatial arrangement of feature mapping on imported training set compounds and their respective experimental activities. Three vital steps: constructive, subtractive and optimization phase direct the aforesaid process [13].
Data analysis
Two theoretical cost calculations; fixed cost and null cost determine the quality of a pharmacophore. On one hand, fixed cost gives off the simplest model robust all the data perfectly, whereas, null cost represents the highest cost of pharmacophore. In null cost, no feature estimates individual activity to be the average of the activity data of the training set compounds. The total cost of any hypothesis ought to be close to the fixed cost and remote from null cost. The cost difference of 40-60 bits amidst fixed cost and null cost signifies 75-90% probability of representing a true correlation in data [14].
The hypotheses for individuals are also assessed on the basis of the error cost, the weight cost and the configuration cost. The entropy of the hypothesis space is measured with the configuration cost. It must be less than a maximum value of 17 and the correlation coefficient must be close to 1.
Pharmacophore validation
rm2 metrics, Fischer’s randomization method, internal test set, and external test set method were used to validate the generated pharmacophore hypothesis.
rm2 metrics: rm2 metrics was calculated in order to check the goodness of generated model. As per recommendations, the ‘average rm2’ and ‘delta rm2’ must be >0.5 and <0.2 [15,16].
Fischer’s randomization method
The correlation between the chemical structure and biological activity was checked and verified by using this method. It generates pharamcophore hypothesis by randomizing the activity data of training set considering same features and the parameters as used to generate original pharmacophore hypothesis. The statistical significance is calculated using formula: Significance=100(1- (1+x/y)), where x represents the total number of hypothesis having a total cost value lower than the original hypothesis, and y represents the total number of HypoGen runs i.e., initial and random runs [17]. 19 random spreadsheets were generated as the confidence level was set to 95%. If the randomized data set results in similar or better cost values, during the generation of pharmacophore, it signifies that the original hypothesis have been generated by chance.
Internal and external test set prediction
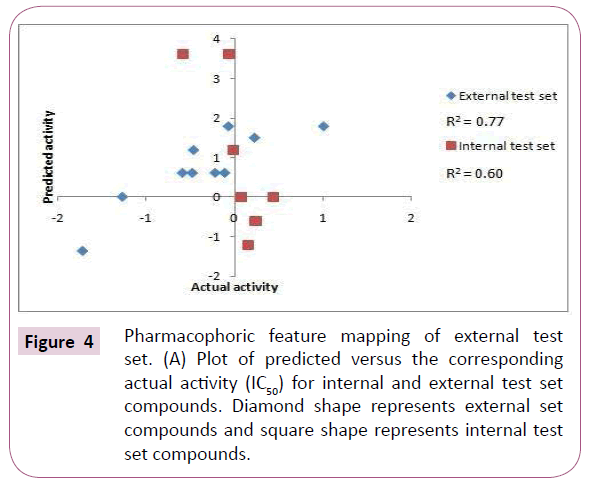
7 internal compounds and 10 external compounds were taken to elucidate the proficiency of the generated pharmacophore to predict the activities of the compounds outside the training set. The correlation coefficient value for internal and external compounds was evaluated [18-20].
Database screening
The database screening from NCI (National Cancer Institute) and Maybridge was employed in order to search novel compounds considering generated pharmacophore as 3-Dimensional query [21]. Retrieved compounds were filtered by applying Lipinski’s rule of five and further sorted on the basis of fit and estimated value.
Molecular docking
The binding modes of the retrieved hits were studied with the help of molecular docking using CDocker program implemented in Discovery Studio (DS) 2.0. Docking studies are based on molecular dynamics algorithm where flexible ligand is docked over rigid receptor in order to attain every possible receptorligand interaction. Crystal structure lanosterol 14α-demethylase (CYP51) was attained from the protein data bank (PDB ID: 3JUS). CDocker interaction energy predicts the binding energies of the protein with retrieved hits [22]. The proximity between the receptor and hit was studied in terms of the hydrophobic, Van Der Waals and hydrogen bond interactions.
Results
Pharmacophore generation
The hypogen module was used to correlate the chemical structures and biological activities of the data set embracing 28 compounds (Table 1). The training set was defined by 21 compounds whereas the test set includes 7 compounds. Removal of three compounds (3h, 3f, and 4c) enduring high error ratio improvised the quality of the model. The pharmacophore model generation was conceded by the training set.

| S No | Name of the compound | R | CYP51 MIC80(µgml-1) | |
|---|---|---|---|---|
| 1 | 3a | H | 1 | |
| 2 | 3b | 4-F | 1 | |
| 3 | 3c | 4-Cl | 0.0625 | |
| 4 | 3d | 2-Cl | 1 | |
| 5 | 3e | 4-Br | 1 | |
| 6 | 3f | 4-CH3 | 0.25 | |
| 7 | 3g | 4-OCH3 | 1 | |
| 8 | 3h | 4-CN | 1 | |
| 9 | 3i | 3-CN | 0.00024 | |
| 10 | 3j | 3-Cl,4-Cl | 4 | |
| 11 | 3k | 4-NO32 | 1 | |
| 12 | 4a |  |
0.25 | |
| 13 | 4b |  |
16 | |
| 14 | 4c |  |
4 | |
| 15 | 4d |  |
0.00024 | |
| 16 | 6a | CH3 | 16 | |
| 17 | 6b | CH2 CH3 | 1 | |
| 18 | 6c | CH2CH2 CH3 | 4 | |
| 19 | 6d | CH2CH2CH2 CH3 | 0.00097 | |
| 20 | 6e | CH2 (CH2)3 CH3 | 0.00097 | |
| 21 | 6f | CH2 (CH2)4CH3 | 0.0039 | |
| 22 | 6g | CH2 (CH2)5CH3 | 4 | |
| 23 | 6h | CH2 CN | 0.25 | |
| 24 | 6i |  |
0.0625 | |
| 25 | 6k |  |
1 | |
| 26 | 6l |  |
1 | |
| 27 | 6m |  |
0.25 | |
| 28 | 6n |  |
0.25 | |
Table 1: Structures and biological activity of Triazole derivatives as CYP51 inhibitors.
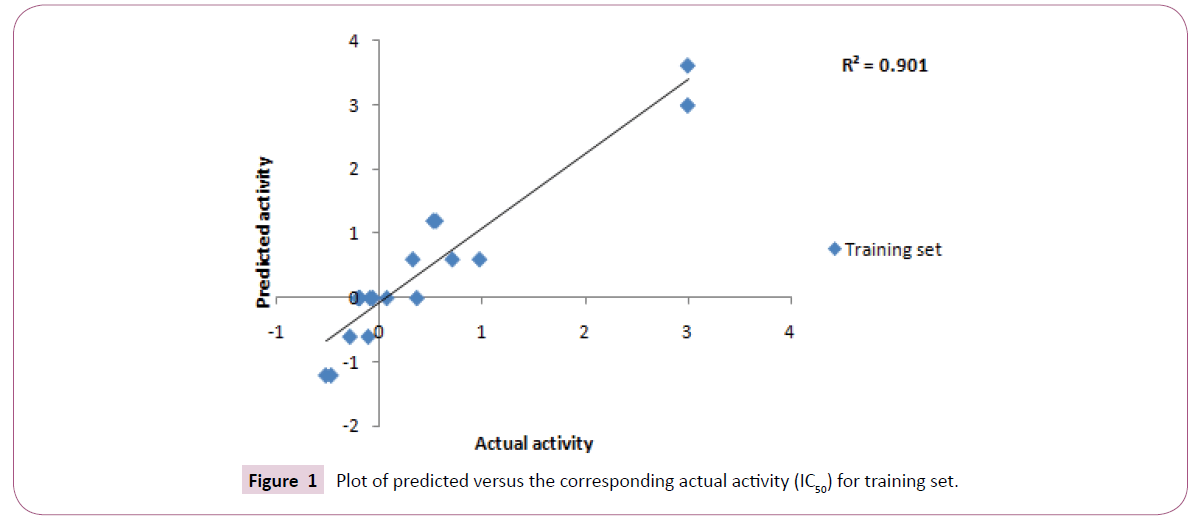
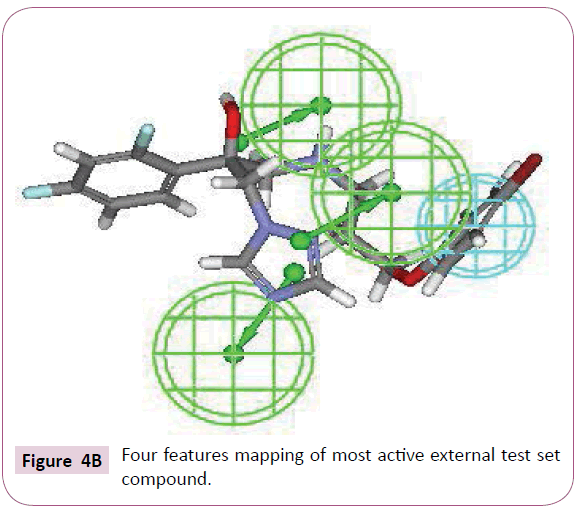
Top ten hypotheses were generated with hydrophobic and HBA as essential chemical features. Significance of statistical parameters was well described through the cost analysis which further validates the pharamacophore. The best hypothesis deals with large cost difference - 33.38), low root mean square - 0.85 and high correlation coefficient - 0.94 (Figure 1). The configuration cost of best hypothesis is 15.82 bits that is not exceeding the recommended value of 17. It might guarantee the intact conformation space sampled all through the pharmacophore generation. All these statistical values specify that the model generated, accounting for all three pharmacophore features: one hydrophobic and three hydrogen bond acceptor (HBA), has good predictive ability (Table 2).
| Name of hits | Estimated value | Fit value | CDocker interaction energy | Structure |
|---|---|---|---|---|
| NSC 1028 | 0.166 | 7.56 | 34.63 |  |
| HTS 00684 | 0.035 | 8.25 | 43.80 |  |
Table 2: Hits retrieved from NCI database as potent CYP51 inhibitors.
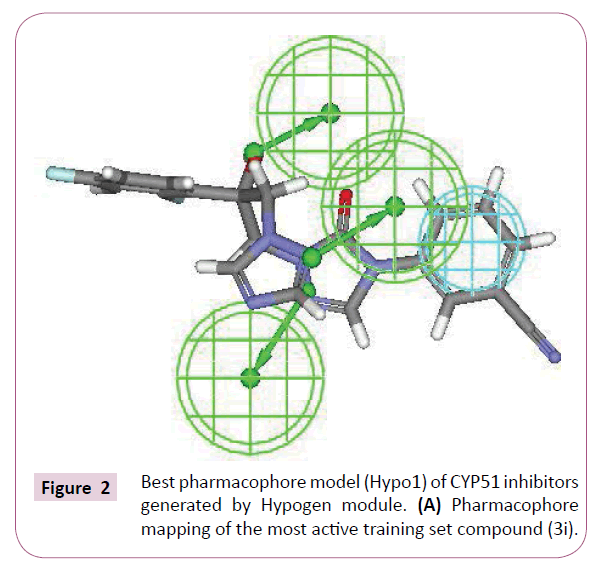
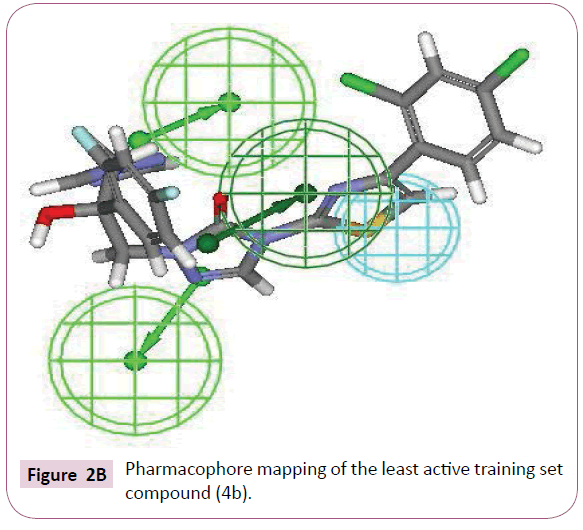
The most active compound (3i) mapped well on all the chemical features showing high fit score of 9.99 (Figure 2A). The pharmacophore superimposition of least active compounds (4b) revealed that it lacks one hydrogen bond acceptor (HBA) feature and showed poor fit value of 6.31 (Figure 2B). The results acquired from aforementioned pharmacophore study appear to validate best hypothesis model to some extent.
Model validation
rm2 metrics: The robustness of the developed model was checked by rm2 metrics. The average rm2 for the training set was found to be 0.78 whereas delta rm2 was observed to be 0.06 which was in relation to the recommended values.
Fischer’s randomization validation
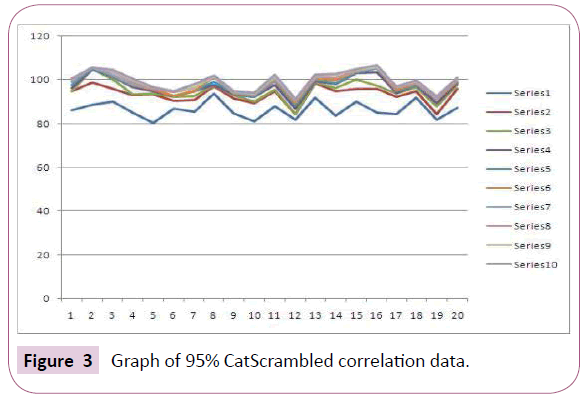
The Fischer’s randomization method further evaluates the statistical relevance of the generated pharmacophore model through verification of the correlation between the chemical structures and the biological activity. The training set has been re-arranged with the aid of the CatScramble program and was used for Hypogen running. CatScramble program generates 19 randomsheets at 95% confidence level. These 19 randomsheets develop hypothesis accounting the same features as used in original pharmacophore hypothesis (Figure 3). The results shows that none of the developed hypothesis has scored a total cost lower than the original hypothesis which indicates the superiority of Hypo 1 statistics. It was clear from this result that the original hypothesis was not generated by chance and has strong confidence to imply a true correlation in the training set.
Internal and external test set validation
The internal and external test set compounds were selected to check the predictive ability of the generated pharmacophore model. The correlation coefficient of 0.61 and 0.70 corresponds to good correlation between actual activity and estimated activity (Figures 4A and 4B).
Virtual screening
The validated pharmacophore model has been used as 3 Dimensional structural query to screen the NCI (National Cancer Institute) and Maybridge databases. As a result, 298 hits from Maybridge and 299 hits from NCI were retrieved. These hits were filtered on the basis of Lipinski’s rule of five. Furthermore, the hits were subsequently mapped over the pharmacophore and those with low fit value were discarded. The low fit value signifies that the centers of the functional groups of the hit are displaced from the centers of the corresponding pharmacophoric features. Finally, 2 hits were retrieved having good fit and low estimated value (Table 3).
| Hypo | Total cost | Cost Difference | RMSD | Correlation | Feature |
|---|---|---|---|---|---|
| 1 | 86.26 | 33.33 | 0.85 | 0.94 | 1HBA, 2HBA lipid, 1HY |
| 2 | 94.68 | 24.91 | 1.20 | 0.89 | 1HBA, 2HBA lipid, 1HY |
| 3 | 94.78 | 24.81 | 1.24 | 0.88 | 1HBA, 2HBA lipid, 1HY |
| 4 | 96.32 | 23.27 | 1.37 | 0.84 | 1HBA, 2HBA lipid, 1HY |
| 5 | 97.56 | 22.03 | 1.33 | 0.87 | 1HBA, 2HBA lipid, 1HY |
| 6 | 99.21 | 20.38 | 1.33 | 0.87 | 1HBA, 2HBA lipid, 1HY |
| 7 | 99.39 | 20.20 | 1.42 | 0.84 | 1HBA, 2HBA lipid, 1HY |
| 8 | 100.08 | 19.51 | 1.46 | 0.83 | 1HBA, 2HBA lipid, 1HY |
| 9 | 100.42 | 19.17 | 1.47 | 0.83 | 1HBA, 2HBA lipid, 1HY |
| 10 | 100.53 | 19.28 | 1.47 | 0.83 | 1HBA, 2HBA lipid, 1HY |
Table 3: Results of the top ten pharmacophore hypotheses generated by the HypoGen algorithm.
Molecular docking
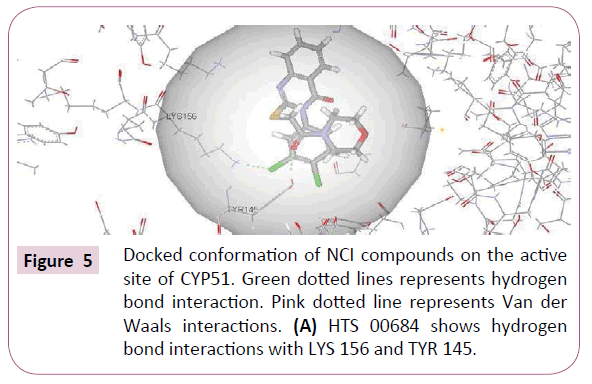
HTS 00684 and NSC 1028 with highest fit value (8.25 and 7.56) and lowest estimated value (0.035 and 0.166) were docked into the active site of CYP51 using CDocker. The crystal structure of CYP51 from the protein data bank (PDB ID: 3JUS) was selected. The binding mode of HTS 00684 with CDocker interaction energy of 43.80 is as shown in (Figure 5A). Chlorine atom of dichlorobenzene moiety showed hydrogen bond interaction with Lys 156 whereas ketonic oxygen of morpholino-2-oxoethylthio showed hydrogen bond interaction with Tyr 145. The chlorine atom of dichlorobenzene has been accommodated within the hydrophobic pocket and interacted with the residues Lys 156, Arg 382, Phe 168 and Leu 117.
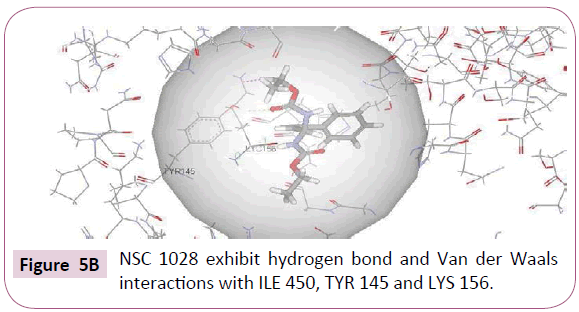
NSC 1028 with CDOCKER interaction energy of 34.63 successfully binds with the active site of CYP51 as shown in (Figure 5B). Specifically, carbonyl oxygen of the carbamate moiety showed hydrogen bond interaction with Tyr 145. Van Der Waals interaction was also observed between methyl group and Lys 156. The benzene group showed hydrophobic interactions with Lys 156, Ile 377, Phe 234, Leu 153, Arg 448 amino acid residues.
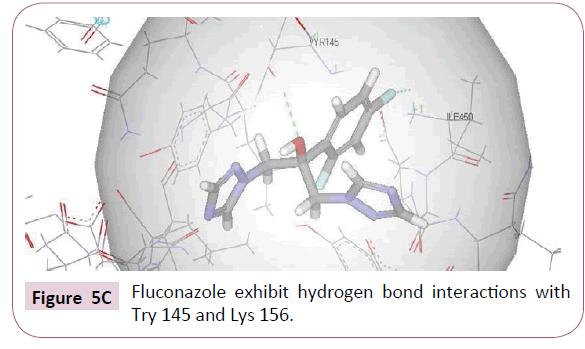
In order to compare the docking results of the test compounds with the standard drug fluconazole was docked into the active site of CYP51. Fluconazole showed three hydrogen bond interactions apart from hydrophobic interactions. 4-fluorine of difluorophenyl showed hydrogen bond interactions with Ile 450 whereas hydroxyl group exhibited hydrogen bond interaction with Tyr 145 (Figure 5C). Benzene ring showed hydrophobic interaction with Ile 450, Arg 448, Lys 160, Lys 156, Lys 157 amino acid residues. It is noticeable that both the test compounds and standard drug exhibit similar kind of interactions with active site amino acids of CYP51.
The studies revealed that hits and standard drugs showed similar kind of interactions with the receptor (CYP51). Furthermore, the novelty of the hits is tested by using pair wise tanimoto similarity indices. HTS 00684 and NSC 1028 showed low tanimoto similarity indices of 0.01 and 0.029 to all structures of established CYP51 confirming their novelty [23].
Discussion
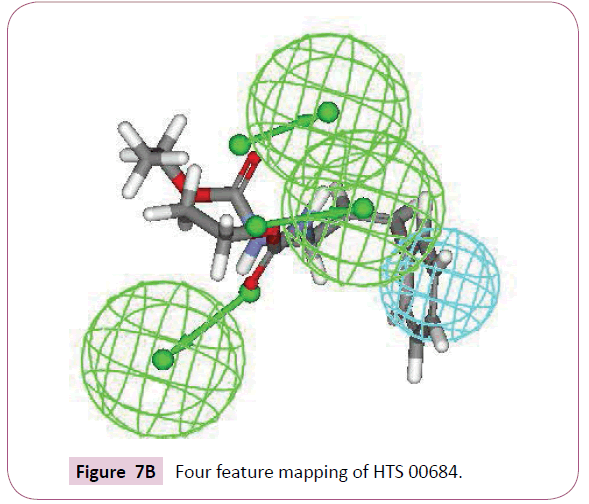
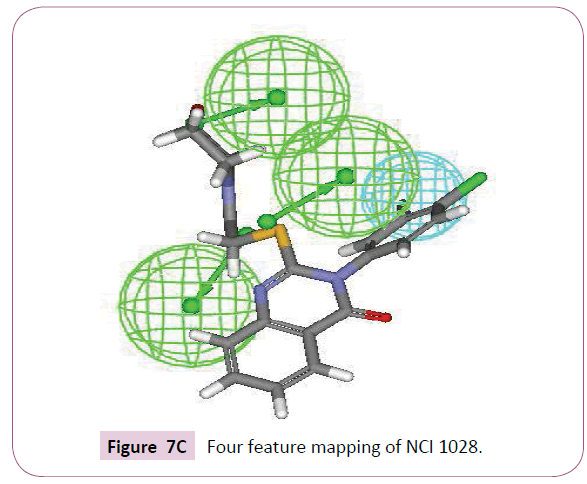
A close inspection of the structures of fluconazole, voriconazole and two lead compounds, reveals that all these compounds contain four common features viz. one hydrophobic group and three hydrogen bond acceptor groups noticeably with almost similar interfeature distances (Figure 6). On comparison of functional groups present in four structures, it has been observed that the difluorobenzene of voriconazole is hydrophobic whereas hydroxyl, azole and fluoropyrimidine groups are forming hydrogen bond within the enzymatic pocket (Figure 7A). Like standard drug voriconazole, one of the identified hit NSC 1028, showed its capability to form hydrogen bond through its carbamate and amine functionality (Figure 7B). The benzene ring of NSC 1028 exhibits hydrophobic interactions. The other hit HTS 00684, due to presence of dichloro benzene functionally exhibits hydrophobic interactions while carbonyl, morpholine and thiol moiety possess hydrogen bond acceptor ability (Figure 7C).
It is evident from the aforementioned analysis that the identified hits possess pharmacophoric features similar to that of fluconazole and voriconazole. Moreover, in fluconazole and voriconazole, presence of substituent at 2nd position could create steric hindrance during its binding within the active site whereas absence of any such group at 2nd position of benzene lowers the activity as testified by the better activity of both the hits in comparison to voriconazole and fluconazole. In context of hydrogen bonding capability of the functional groups, it is noteworthy that the fluconazole and voriconazole contains sp2 nitrogen and sp3 hydroxyl whereas NSC 1028 attain one sp3 oxygen and two sp2 carbonyl group and HTS 00684, contains sp2 carbonyl and thiol group. Difference in the acidity (pKHA) value which means the strength of hydrogen bond acceptor [24] clearly supports the high activity (low estimated and high fit value) of NSC 1028 and HTS 00684. It is a well-known fact that azoles bind to CYP51 enzyme through a coordinate bond with Fe (3+ low spin state) whereas in case of NSC 1028 (Figure 8A), nitrogen adjacent to ester group forms a coordinate bond with Fe (3+ low spin). On the other hand, in HTS 00684 (Figure 8B), coordination bond was observed between nitrogen of morpholine moiety and Fe (3+ low spin) confirming the CYP51 inhibitory activity of the identified hits.
Conclusion
Our study illustrates the identification of two potential lead compounds which could be further evaluated using in silico and in vitro techniques.
Acknowledgements
Computational resources were provided by Banasthali University, and the authors thank Vice Chancellor for providing necessary facilities.
References
- Diekema DJ, Pfaller MA (2004) Nosocomial Candidemia: An Ounce of Prevention Is Better Than a Pound of Cure. Infection Control & Hospital Epidemiology 25:624-626.
- Rees JR, Pinner RW, Hajjeh RA, Brandt MR, Reingold AL (1998) The epidemiological features of invasive mycotic infections in San Francisco Bay area, 1992-1993: results of population-based laboratoryactive surveillance.Clinical Infectious Diseases 27:1138-1147.
- Ekins S, Mankowski DC, Hoover DJ, Lawton MP, Treadway JL, et al. (2007) Three-dimensional quantitative structure-activity relationship analysis of human CYP51 inhibitors. Drug Metabolism and Disposition35:493-500.
- Ghannoum MA, Rice LB (1999)Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clinical Microbiology Reviews 12: 501-517.
- Rodriguez RJ, Low C, Bottema CDK, Parks LW (1985) Multiple functions for sterols in Saccharomyces cerevisiae.BiochimicaetBiophysicaActa837: 336-343.
- Aoyama Y, Yoshida Y, Sonoda Y, Sato Y(1989) Deformylation of 32-oxo-24,25-dihydrolanosterol by the purified cytochromeP-450 14DM (lanosterol 14α-demethylase) from yeast evidence confirming the intermediate step of lanosterol 14α-demethylation.Journal of Biological Chemistry 264: 18502-18505.
- Alexander K, Akhtar M, Boar RB, McGhie J, Barton FD,et al. (1972) The removal of the 32-carbon atom as formic acid in cholesterol biosynthesis. Journal of the Chemical Society, Chemical Communications.pp: 383-385.
- HeQQ, LiK, Cao YB, DongHW, ZhaoLH, et al. (2007)Design, synthesis and molecular docking studies of novel triazole antifungal compoundChinese Chemical Letters 18:663-666.
- Chakrabarti A (2011) Drug resistance in fungi- an emerging problem. Regional Health Forum15:97-103.
- Valasani KR, Vangavaragu JR, Day VW, Yan SS (2014) Structure based design, synthesis, pharmacophore modeling, virtual screening, and molecular docking studies for identification of novel cyclophilin D inhibitors.Journal of Chemical Infromatics and Modeling 54:902-912.
- Jiang YY, Zhang J, Zou Y, Chai X, et al. (2011) Design, synthesis and antifungal evaluation of 1-(2-(2,4-difluorophenyl)-2-hydroxy-3-(1H-1,2,4-triazol-1-yl)propyl)-1H-1,2,4-triazol-5(4H)-one. European Journal of Medicinal Chemistry46: 3135-3141.
- Chen IJ, Foloppe N (2008) Conformational sampling of druglike molecules with MOE and catalyst: implications for pharmacophoremodeling and virtual screening. Journal of Chemical Infromatics and Modeling48:1773-1791.
- Mason JS, Good AC, Martin EJ (2001) 3D pharmacophores in drug discovery. Current Pharmaceutical Design7:567-597.
- Arooj M, Thangapandian S, John S, Hwang S, Park JK, et al. (2011) 3D QSAR pharmacophore modeling, in silico screening, and density functional theory (DFT) approaches for identification of human chymase inhibitors.International Journal of Molecular Sciences 12:9236-9264.
- RoyK, ChakrabortyP, MitraI, OjhaPK, KarS, et al. (2013)Some case studies on application of "(rm)2" metrics for judging quality of quantitative structure-activity relationship predictions: emphasis on scaling of response data.Journal of Computational Chemistry 34:1071-1082.
- MitraI, SahaA, RoyK(2013)Predictive Modeling of Antioxidant Coumarin Derivatives Using Multiple Approaches: Descriptor-Based QSAR, 3D-Pharmacophore Mapping, and HQSAR. ScientiaPharmaceutica 81:57-80.
- BharathamN, BharathamK, LeeKW(2007)Pharmacophore identification and virtual screening for methionyl-tRNAsynthetase inhibitors. Journal of Molecular Graphics and Modeling25:813-823.
- Sheng C,Zhang W, Ji H, Zhang M, Song Y, et al. (2006) Structure-based optimization of azole antifungal agents by CoMFA, CoMSIA and molecular docking. Journal Medicinal Chemistry 49:2512-2525.
- Bhattacharjee A, Mylliemngap BJ, Velmurugan D (2012)3D-QSAR studies on fluroquinolones derivatives as inhibitors for tuberculosis. Bioinformation 8:381-387.
- Mehta H, Khokra SL, Arora K, Kaushik P (2012)Pharmacophore mapping and 3D-QSAR analysis of Staphylococcus AureusSortase a inhibitors. Der PharmaChemica 4:1776-1784.
- Xiang M, Lei K, Fan W, Lin Y, He G, et al. (2013) Insilico identification of EGFR-T790M inhibitors with novel scaffolds: start with extraction of common features. Drug Design Development and Therapy7:789-839.
- Shin HL, Josephine WW, Hsuan LL, Jian HZ, Kung TL, et al. (2011) The discovery of potential acetylcholinesterase inhibitors: A combination of pharmacophore modeling, virtual screening, and molecular docking studies. Journal of Biomedical Science 18:8.
- Shen MY, Yu H, Li Y, Li P, Pan P, et al. (2013) Discovery of Rho-kinase inhibitors by docking-based virtual screening. Molecular BioSystems 9:1511-1521.
- Laurence C, Berthelot M (2000) Observation on the Strength of Hydrogen Bonding. Perspectives in Drug Discovery and Design 18:39-60.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences