Inter-helical Electrostatic Interaction and Rotameric Signatures in Activated Sphingosine-1-Phosphate Receptor 1
Olaposi I Omotuyi, Hiroshi Ueda
Olaposi I Omotuyi1,2 Hiroshi Ueda1,2*
Department of Pharmacology and Therapeutic Innovation, Graduate School of Biomedical Sciences, Nagasaki University, Japan
Center for Drug Discovery and Therapeutic Innovation, Nagasaki University, Japan
- *Corresponding Author:
- Hiroshi Ueda
Department of Pharmacology and Therapeutic Innovation
Graduate School of Biomedical Sciences, Nagasaki University
Japan
Tel: +81958192421
E-mail: ueda@nagasaki-u.ac.jp
Received date: June 09, 2015; Accepted date: July 13, 2015; Published date: July 16, 2016
Citation: Omotuyi OI, Ueda H. Inter-helical Electrostatic Interaction and Rotameric Signatures in Activated Sphingosine-1- Phosphate Receptor 1. Chem Inform. 2015, 1:1.
Abstract
Sphingosine-1 phosphate (S1P) receptor 1 (S1PR) is one of the receptors responsible for initiating intracellular signal transduction in response to extracellular signals encoded in Sphingosine-1 phosphate (S1P) and other related agonists. Using microsecond molecular dynamics simulation, rotameric (χ2) changes in the aromatic amino acids lining ligand-binding pocket, inter-helical electrostatic interaction, interaction between ligands (S1P and ML056) and selected extracellular regions of S1P have been studied. The data presented here strongly suggested that S1P-bound S1PR structure became active based on NPxxYmotif rmsd and dissociation of TM3/TM6 ionic lock as early as 300 ns while the apo- and ML056-bound S1PR were trapped in semi-active and inactive states respectively. Tyrosine 29 evolved agonist-dependent rotameric distribution while tryptophan 117 (W3.25), phenylalanine 210 (F5.47), tryptophan 269 (W6.48) and phenylalanine 273 (F6.52) exhibited activation-type signatures. Furthermore, while activation promoted TM1/TM4, TM2/TM7 and TM4/TM6 engagement, prior electrostatic engagements in TM3/TM4, TM3/TM6 and TM3/TM7 were dissolved. N-terminal-heptahelical bundle interaction is also compromised in inactive S1PR possibly due to reduced engagement of N-terminal residue such as lysine 34 and lysine 46. Ultimately, S1PR activation by class I agonist followed classical GPCR activation paradigm.
Keywords
Sphingosine-1 phosphate (S1P) receptor; Rotameric signatures; Electrostatic interaction
Introduction
Sphingosine-1-phosphate receptor 1 (S1PR, also referred to as endothelial differentiation gene 1 (S1PR)) is one of the 5 evolutionarily related G-protein-coupled receptors (GPCRs), which confer bioactivity on sphingosine 1-phosphate (S1P) [1]. Dysfunctional S1P/S1PR signaling has been implicated in experimental autoimmune encephalomyelitis (EAE) and other autoimmune neuro-inflammation phenotypes [2], thus lending strong evidence for the approval (FDA-approved) of fingolimod (FTY720) as an immuno-modulating drug for the management of relapsing-remitting multiple sclerosis via S1PR mechanism [3]. Toward deeper understanding of ligand-S1PR interaction, two x-ray structures (PDB IDs: 3V2W (3.35 Å) and 3V2Y (2.80 Å)) of S1PR in complex with an antagonist (ML 056) have been deposited into the protein data bank repository [4]. These structures did provide the starting structure for the first detailed molecular dynamics simulation study of ligand binding and receptor activation mechanisms [5, 6]. A major highlight from the crystal structure of S1PR is that interaction between aromatic residues lining the ligand pocket and S1PR-active ligands drives ligand selectivity, activation and antagonism mechanisms [4]. Further mechanistic insight provided by microsecond molecular dynamics (MD) simulation study showed that when ML056 is replaced with S1P within the orthosteric site, movement of S1P tail carbon atoms is coupled to flipping of tryptophan 269(TM6.48) which resonates through proximal phenylalanine 265(TM6.44) thus, forcing a similar dihedral change. Ultimately, a transmission switch is initiated from these local events which propagates through asparagine 63(1.50), aspartate 91(2.50), serine 304(7.46) and asparagine 307(7.49) causing influx of water molecules and large movement in transmembrane helices (the cytoplasmic region) [5, 6]. In this study, we re-investigated the dynamics of Apo-S1P1, ML056-S1P1 (antagonist-bound) and S1P-S1P1 (agonist-bound) at 1.5 μs (doubling the previously reported production phase time of 700 ns) [5] in order to provide further insight into S1P1 activation mechanism with a focus on identifying aromatic dihedral signatures present in the N-terminal region which relays ligand-dependent (and independent) actions through the aromatic residues lining the orthosteric pocket of S1PR, and how activation of the receptor re-dictates electrostatic interaction between transmembrane (TM) helices.
Materials and Methods
Starting Structures
The crystal structure of S1P receptor resolved at 2.80 Å (PDB ID: 3V2Y) was retrieved from the protein data bank (PDB) to prepare the starting co-ordinates. The missing intracellular loop (ICL)-II (residues 149-155) and ICL-III (residues 232-244) in the crystal structure was replaced using loop-modelling suites in MODELLER (version 9v13) and ROSETTA as previously documented [5]. In both cases, the lowest DOPE-scored loop was selected from the pool of 5000 loop conformations generated using MODELLER software, was piped into the ROSETTA for refinement as reported. To make the starting apo-S1PR coordinate, C-terminus residues beyond helix H8 region was removed. To generate ML056-S1PR coordinate, apo-S1PR and the x-ray structure (3V2Y) coordinates were superimposed using align module in PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrodinger, LLC.), followed by the addition of ML056 coordinate from the x-ray structure to apo-S1PR coordinate. To generate starting S1P-S1PR coordinate, 2D coordinate of S1P (C18-Sphingosine 1-phosphate, CID 5283560) obtained from PUBCHEM database [7] converted into a low-energy 3D coordinate using LowModeMD conformational search module in Molecular Operating Environment (MOE, Molecular Operating Environment (MOE), 2013.08; Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2015). The 3D-coordinate was aligned with ML056 (reference, from ML056-S1PR) coordinate using LigAlign [8] plugin in PyMOL, the new S1P coordinate was added to apo-S1PR.
Biosystems setup
Orientation (along the membrane normal) of the biosystems was performed using the PPM server (opm.phar.umich.edu/server. php) [9]. Each of the oriented biosystem was inserted into a preequilibrated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, 68 lipids per leaflet) bilayer using CHARMM-GUI webserver (www.charmm-gui.org) [10]. Ligand (S1P and ML056) parametization was performed using ParamChem service (https://cgenff.paramchem.org) as implemented on CHARMMGUI webserver. The biosystems were solvated in TIP3P explicit water model and neutralized with Na=/CL- (0.15 M).
Molecular dynamics (MD) simulation
All molecular dynamics simulation (GROMACS, ver. 4.6) [11] was performed using CHARMM36 force field [12]. During equilibration, the biosystems were subjected to constant pressure and temperature (NPT; 310K, 1 bar) conditions as implemented in Berendsen temperature and pressure coupling algorithms [13] as implemented in GROMACS. Van der Waals interactions were estimated at 10 Å, long-range electrostatic interactions were computed using particle mesh Ewald (PME) summation scheme [14] while equation of atomic motion was integrated using the leap-frog algorithm [15] at 2 fs time step for a total time of 100 ns with positional restraints imposed on the heavy atoms in all directions. To get two (n=2) starting coordinates for the production phase, an initial 1 ns unrestrained simulation was performed on each biosystem and the co-ordinates at 500 ps and 800 ps were harvested from each biosystem for long (1.5 μs) production phase simulation.
Data Analysis
For data analysis, in built analysis tool kit in GROMACS [11] was utilized. In this study, g_mindist module (for interatomic minimum distance analysis), g_dist module (for estimating the center of mass distance between two groups), g_angle (for estimating the chi2 (χ2) dihedral angles) and g_sham (data set for 3D energy landscape plots) were used. Graphs were generated using MATHEMATICA (Wolfram Research, Inc, ver. 10.0, Champaign, Illinois, 2014) statistical software (3D plots) and graphpad prism (GraphPad Software, San Diego California USA, www.graphpad. com) was used for generating line plots and bar charts.
Results and Discussion
Structural basis for ML056 and S1P actions on S1PR
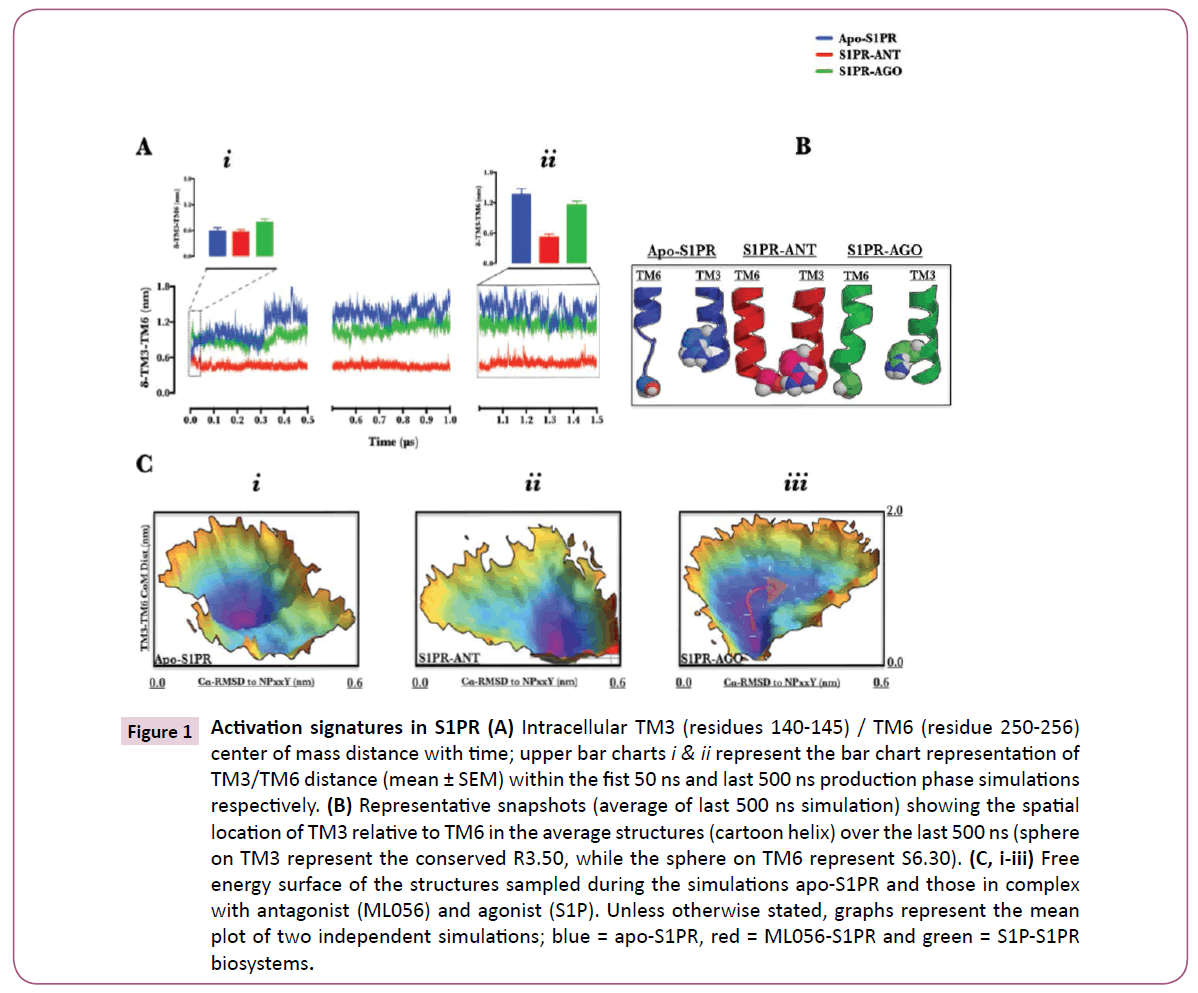
Physiologically, GPCRs exist in multiple but dynamic conformational states spanning inactive and active spectrum [16]. GPCR-modulatory compounds are known to stabilize specific conformations or ease the transition between conformations [17]. Sphingosine-1-phosphate receptor 1 (S1PR) is a member of GPCR family of proteins and the biological target of Sphingosine-1-phosphate (S1P) and fingolimod [1, 3, 4, 6, 18]. In order to further understand the mechanism underlying S1PR activation, first we investigated the conformations stabilized by Sphingosine-1-phosphate and ML056 [4] in comparison with the apo-S1PR biosystem. Whilst the starting structures were essentially inactive as informed by TM3/TM6 center of mass distance ≈ 0.6 depicting an ionic lock [19] (Fig. 1A, upper bar chart (I)), at ≈ 300 ns (Fig. 1A), apo-S1PR and sphingosine-1- phosphate-bound S1PR began to evolve active conformations with broken TM3/TM6 ionic lock (transmembrane (TM)3/TM6 distance > 0.6) which is a key activation signature; [5] the broken ionic lock was maintained till 1.5 μs (Fig. 1A, upper bar chart (II)). Indeed, the average structures of protein within the last 500 ns of the simulations showed that TM3 and TM6 were dissociated in apo- and sphingosine-1-phosphate-bound but not ML056- bound S1PR Figure 1B. TM3/TM6 ionic lock dissociation alone does not fully explain activation episode in GPCRs; another key activation signature is found in the conserved NPxxY motif, where water tunneling into activated GPCR is under investigation [5, 20, 21]. By projecting the data collated from TM3/TM6 distance and root mean square deviation of NPxxY motif to an inactive conformation along the 3-D free energy landscape (Figure 1C, i, ii & iii), we provided further insight into metastable conformational states sampled by S1PR biosystems during the simulation. The result showed each set-up sampled different conformations during the simulation. In apo-S1PR (Figure 1C, i), sampled metastable conformations have broken TM3/TM6 ionic lock but the NPxxY motif was essentially inactive (Cα-RMSD to NPxxY ≤ 0.3 nm) whereas, ML056 bound EDG1 (Figure 1C, ii), which paradoxically had its TM3/TM6 trapped in an inactive ionic lock did evolve active NPxxY motif conformation (Cα-RMSD to NPxxY > 0.5 nm). S1P-bound S1PR (Figure 1C, iii) surprisingly showed a dynamic flow of structures moving from inactive NPxxY motif conformation to active conformation (Cα-RMSD to NPxxY > 0.4 nm), concomitantly with broken TM3/TM6 ionic lock by removing energy barriers (ΔG ≈ 0 Kj/mol, see arrow). Clearly, for successful activation of S1PR, active NPxxY motif conformation (Cα-RMSD to NPxxY > 0.4 nm) must be coupled with increase in TM3/TM6 center of mass distance and neither of the two events alone may fully activate S1PR. Indeed, in rhodopsin, the conformational changes occurring at the NPxxY motif allows TM7 to insert into the 3D-space previously occupied by TM6 (when ionic lock was formed with TM3) thereby preventing the re-formation of the ionic lock and ultimately stabilizing the activated rhodopsin [22]. Furthermore, some inactive GPCRs conformations have been crystallized with broken TM3/TM6 ionic lock [23] similarly, in a series of experiments on Histamine H4 receptor, mutants that lack ionic lock-forming capacity (R6.30A) did not promote G-protein activation. In similar fashion, ionic lock-promoting mutant (R6.30E) did not alter its constitutive activity; taken together, the authors concluded that reversible TM3/TM6 ionic lock formation may not be a general requirement for all class A GPCRs [24]. In a recent review[25], the authors pointed the absence of ionic lock in β2-AR/carazolol complex, and in subsets of A2AAR, β1AR, and D3R [23, 26]. Therefore, data presented here fully demonstrated that apo- and ML056-bound S1PR sufficiently sampled semiactive (intermediate, broken ionic lock without accompanying NPxxY motif conformational change) and inactive (activationtype NPxxY with intact ionic lock, signature of an inverse agonist) states respectively while S1P-S1PR demonstrated active-state signatures. Semi-active conformation sampled by apo-S1PR may provide a plausible explanation for the basal activity (agonistindepedent activation) reported in S1PR [27] just as the data provided further insight into the structural basis for ML056 and S1P antagonistic and agonistic actions respectively.
Figure 1: Activation signatures in S1PR (A) Intracellular TM3 (residues 140-145) / TM6 (residue 250-256) center of mass distance with time; upper bar charts i & ii represent the bar chart representation of TM3/TM6 distance (mean ± SEM) within the fist 50 ns and last 500 ns production phase simulations respectively. (B) Representative snapshots (average of last 500 ns simulation) showing the spatial location of TM3 relative to TM6 in the average structures (cartoon helix) over the last 500 ns (sphere on TM3 represent the conserved R3.50, while the sphere on TM6 represent S6.30). (C, i-iii) Free energy surface of the structures sampled during the simulations apo-S1PR and those in complex with antagonist (ML056) and agonist (S1P). Unless otherwise stated, graphs represent the mean plot of two independent simulations; blue = apo-S1PR, red = ML056-S1PR and green = S1P-S1PR biosystems.
Rotameric signatures in aromatic amino acids lining S1P1 orthosteric pocket
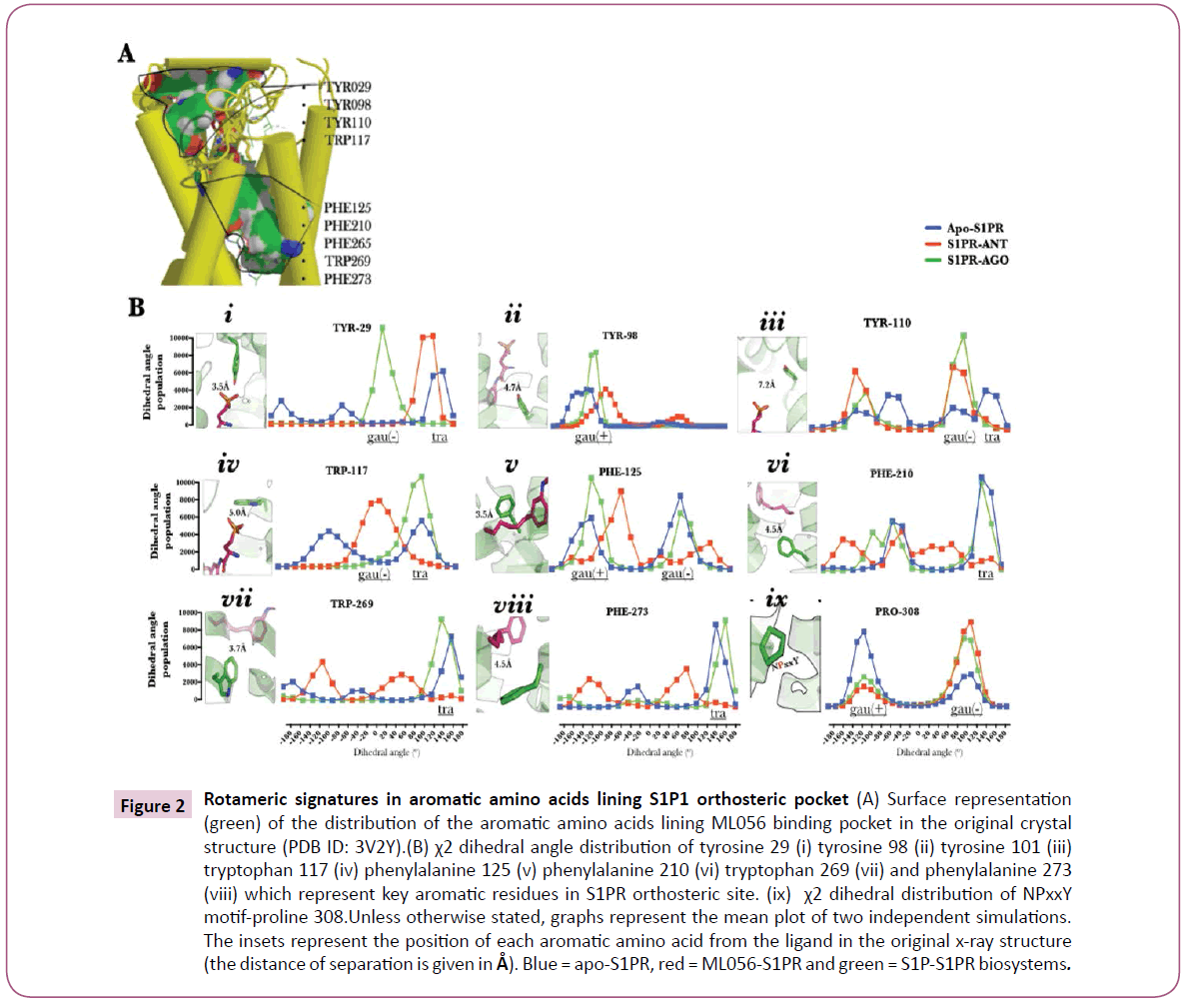
S1PR activation signatures evoked by S1P can only be explained by interaction between residues lining the orthosteric pocket and S1P. Undoubtedly, discretely localized ligand-residue interactions must be systematically ordered into robust topological changes in the heptahelical bundle, a key driver in G-protein coupling and activation process [1, 22]. In this study, a detailed study of interaction between S1PR-active ligands (agonist and antagonist) and the aromatic residues proximal to ML056 in the original crystal structure (PDB ID 3V2Y) [4] has been investigated in terms of side chain dihedral (χ2) changes Figure 2A. In S1P-bound biosystem, N-terminal tyrosine 29 (TYR-29, occurring at 3.5 Å from ML056 phosphate-head group, inset, (Figure 2B, i) assumed a gauche (-) dihedral (0≤ χ2≤120) while in other biosystems (ML056-S1PR and apo-S1PR), trans dihedral angle 120≤χ2<180; −180≤χ2<−120, (Figure 2B, i) was preferentially sampled. Although in previous study, a post-equilibration stable interaction was observed between this residue and the ligands investigated [5], however, the rotameric effects of the ligands were distinct.
Transmembrane-2 tyrosine 98 (Y2.57) located at 4.7 Å from the head group of ML056 inset, (Figure 2B, ii) sampled gauche (-) dihedral (-120≤χ2<0) independent of biosystem set up but at higher population in agonist bound S1PR compared with the other biosystems (Figure 2B, ii). The restraint on the dihedral space sampled by tyrosine 98 in ML056 bound-S1PR may be explained by the presence of hydrogen bond between the carbonyl group of ML056 and the phenolic group of the residue and in apo-state, formation of a hydrogen bond with serine 304 (7.46) [5].
Dihedral changes in tyrosine 110 (ECL-I residue located at 7.4 Å from the head group of ML056, inset, (Figure 2B, iii) provided the first ligand-dependent dihedral event during observed in this study; here, agonist and antagonist-bound biosystems preferentially sampled gauche (-) dihedral and gauche (+) dihedral. When this data is interpreted within the context of previous findings that ECL-1 occludes the orthosteric site [4], tyrosine 110 may therefore represent one of the hypothetical ligand-sensors, which drives structural compactness of ECL-1 and the N-terminal.
Based on the observation that S1P-bound S1PR evolved activation-type molecular and structural signatures [5] and the apo-S1PR is in an intermediate state, then activation-type (distinct pattern observable in agonist- and apo-S1PR) dihedral change was assumed to be observed in tryptophan 117 (W3.25, located 5.0 Å from the head group of ML056, inset, Figure 2B, iv) where sampled trans (120≤χ2<180) dihedral space was sampled in apo-S1PR and agonist-bound S1PR biosystems (Figure 2B, iv), phenylalanine 125 (F3.33, located 3.5 Å from the tail carbon atoms of ML056, inset, Figure 2B, v) also sampled activationtype gauche (-) and gauche (+) dihedral space (Figure 2B, v), phenylalanine 210 (5.47, located 4.5 Å from the tail carbon atoms of ML056, inset, Figure 2B, vi), tryptophan 269 (W6.48, located 3.7 Å from the tail carbon atoms of ML056, inset, Figure 2B, vii), and phenylalanine 273 (F6.52, located 4.5 Å from the tail carbon atoms of ML056, inset, Figure 2B, viii) (Figure 2B, vi, vii & viii). First, dihedral changes in phenylalanine 125, 210, and 273 may have been mediated by hydrophobic interaction between the aromatic nuclei of the residues and the hydrophilic tail of S1P [5], which may be, enhanced in aromatic-moiety rich class II S1PR agonists [28] which do not depend on the traditional arginine 120 (3.28)/glutamate 121 (3.29) for receptor activation as class I agonists [4, 5, 27, 28]. Secondly and more importantly, the observed rotameric distribution pattern of tryptophan 269 in agonist bound biosystem as presented here is similar to χ2 angle associated with transmission switch during S1PR activation reported in previous study [5] and other class-A GPCRs [5, 22, 29, 30].
Figure 2: Rotameric signatures in aromatic amino acids lining S1P1 orthosteric pocket (A) Surface representation (green) of the distribution of the aromatic amino acids lining ML056 binding pocket in the original crystal structure (PDB ID: 3V2Y).(B) χ2 dihedral angle distribution of tyrosine 29 (i) tyrosine 98 (ii) tyrosine 101 (iii) tryptophan 117 (iv) phenylalanine 125 (v) phenylalanine 210 (vi) tryptophan 269 (vii) and phenylalanine 273 (viii) which represent key aromatic residues in S1PR orthosteric site. (ix) χ2 dihedral distribution of NPxxY motif-proline 308.Unless otherwise stated, graphs represent the mean plot of two independent simulations. The insets represent the position of each aromatic amino acid from the ligand in the original x-ray structure (the distance of separation is given in Å). Blue = apo-S1PR, red = ML056-S1PR and green = S1P-S1PR biosystems.
Perhaps, the most interesting result yet in the activation-type χ2 dihedral series was observed in the proline (proline 308, P7.50) residue of NPxxY motif. In most GPCRs [5, 22, 29], rotameric changes in tryptophan 269 (transmission switch) occur mutually inclusive of large structural rearrangement in TM7 mediated by the proline component of NPxxY motif. In this study, proline 308 in agonist- and antagonist-bound S1PR preferentially sampled gauche (+) χ2 (-120≤χ2<0) dihedral space while trapped in gauche (-) χ2 (0≤χ2<120) dihedral space in semi-active S1PR (apo-state); as observed in the 3D-free energy surface plot. This data indicated that NPxxY kink might not be coupled with the movement of tyrosine 311 (307-NPxxY-311) (Y7.53) into the heptahelical bundle (required for maintaining GPCRs in an active state). Similarly, the data lend a valuable credence to the proposition that GPCRs conformation may not be explained in two state (ON/OFF for activated and inactivate states), rather, a more acceptable multi-state “molecular rheostats-like” state in continuum conformation dynamics separated in space and time by energy has been proposed [22, 25].
Energetics of transmembrane helix interaction and ligand-S1PR binding
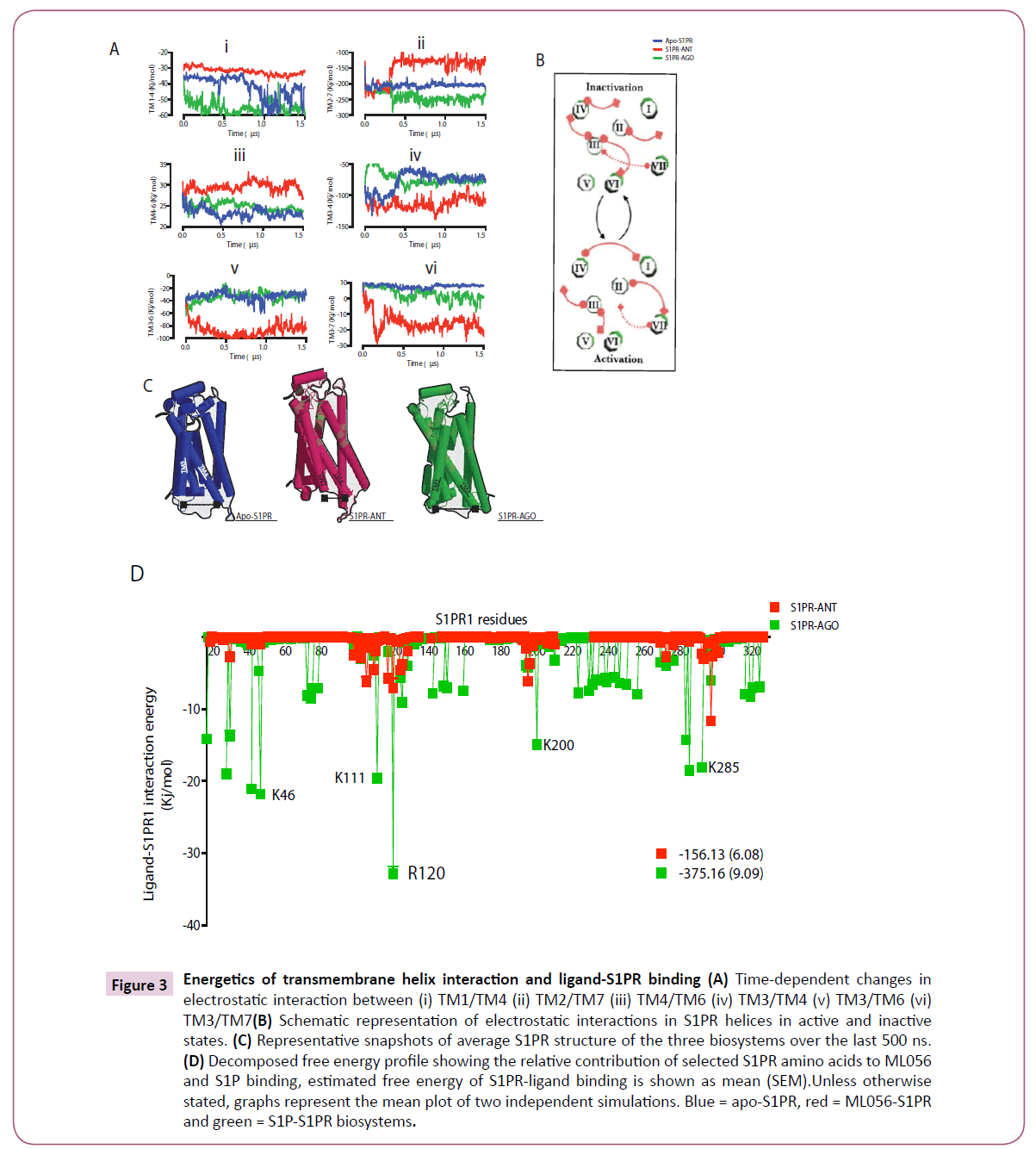
Ultimately, the local events at GPCR transmembrane helices sum up to cause large changes in the overall receptor topology thereby driving activation and inaction [4, 5, 20, 22, 25, 29]. Since earlier in this study, we have provided some evidence, which suggested that apo- and agonist bound S1PR showed activationtype structural arrangement and dihedral signatures, next, we sought to elucidate the energy of interaction between specific TM helices in the distinct conformational states identified in this study (active (agonist bound), semi-active (intermediate, apostate) and inactive (antagonist bound) states). Data presented here suggested that during S1PR activation (semi-activation and full-activation), electrostatic interaction is essential between TM1 (residue 44-70) and TM4 (residue 170-177); such interaction seems lost in inactivated S1PR (Figure 3A, i). In similar fashion, the intensity of TM2 (residue 83-102) / TM7 (residue 293- 312) interaction showed correlation with receptor activation. Interesting, starting at ≈ 300 ns, the three biosystems began to evolve a distinct pattern (Fig. 3A, ii) as fully active (agonist bound) S1PR interaction energy ≈ -270 Kj/mol till the end of the simulation, the interaction energy of the semi active biosystem was tightly maintained at -200 Kj/mol while the antagonist bound biosystem evolved the least intense interaction with the average energy value of -110 Kj/mol (Figure 3A, iii). Additionally, a weak interaction between TM4/TM6 (residue 252-278) may also contribute to S1PR activation (Figure 3A, iii). In contrast, repulsion in TM3/TM4 (-75 vs. -120 Kj/mol for active and inactive respectively, Figure 3A, iv), TM3 (residue 117-144)/TM6 (-40 vs. -100 Kj/mol for active and inactive respectively, Figure 3A, v) and TM3/TM7 (10 vs. -12 Kj/mol for active and inactive respectively, Figure 3A, vi) may be required for S1PR activation. Whereas TM3/ TM6 repulsion is the most commonly documented structural for activation in almost all known GPCRs [4, 5, 20, 20, 22, 25, 29,31], this study reports that electrostatic interaction was also lost between TM3/TM4 and TM3/TM7 during S1PR activation. It is interesting to note that disruption in salt-bridge (electrostatic interaction) between TM3 and TM7 reportedly cause constitutive activation in agreement with our findings[31, 32] and that large repulsion between these helices are counter-balanced with attractions in other TM helices as observed n TM2/TM7 and TM4/TM6.
The contribution of transmembrane interaction to S1PR activation is presented in the schematic diagram (Figure 3B) and the average structure of the three biosystems within the last 500 ns showed a large displacement between TM3 and TM4 (Figure 3C). In terms of binding affinity, sphingosine-1-phosphate showed two-fold higher energy of interaction with S1PR compared with ML056 (-375.16 vs. -156.13 Kj/mol) and the difference in affinity may be traced to its higher interaction with lysine 46 (K1.33), lysine 111 (ECL-II), arginine 120 (R3.28), lysine 200 (K5.37) and lysine 285 (ECL-III) (Figure 3D).
Figure 3: Energetics of transmembrane helix interaction and ligand-S1PR binding (A) Time-dependent changes in electrostatic interaction between (i) TM1/TM4 (ii) TM2/TM7 (iii) TM4/TM6 (iv) TM3/TM4 (v) TM3/TM6 (vi) TM3/TM7(B) Schematic representation of electrostatic interactions in S1PR helices in active and inactive states. (C) Representative snapshots of average S1PR structure of the three biosystems over the last 500 ns. (D) Decomposed free energy profile showing the relative contribution of selected S1PR amino acids to ML056 and S1P binding, estimated free energy of S1PR-ligand binding is shown as mean (SEM).Unless otherwise stated, graphs represent the mean plot of two independent simulations. Blue = apo-S1PR, red = ML056-S1PR and green = S1P-S1PR biosystems.
Ligand binding causes heptahelical bundle packing with extracellular loops and the N-terminal region
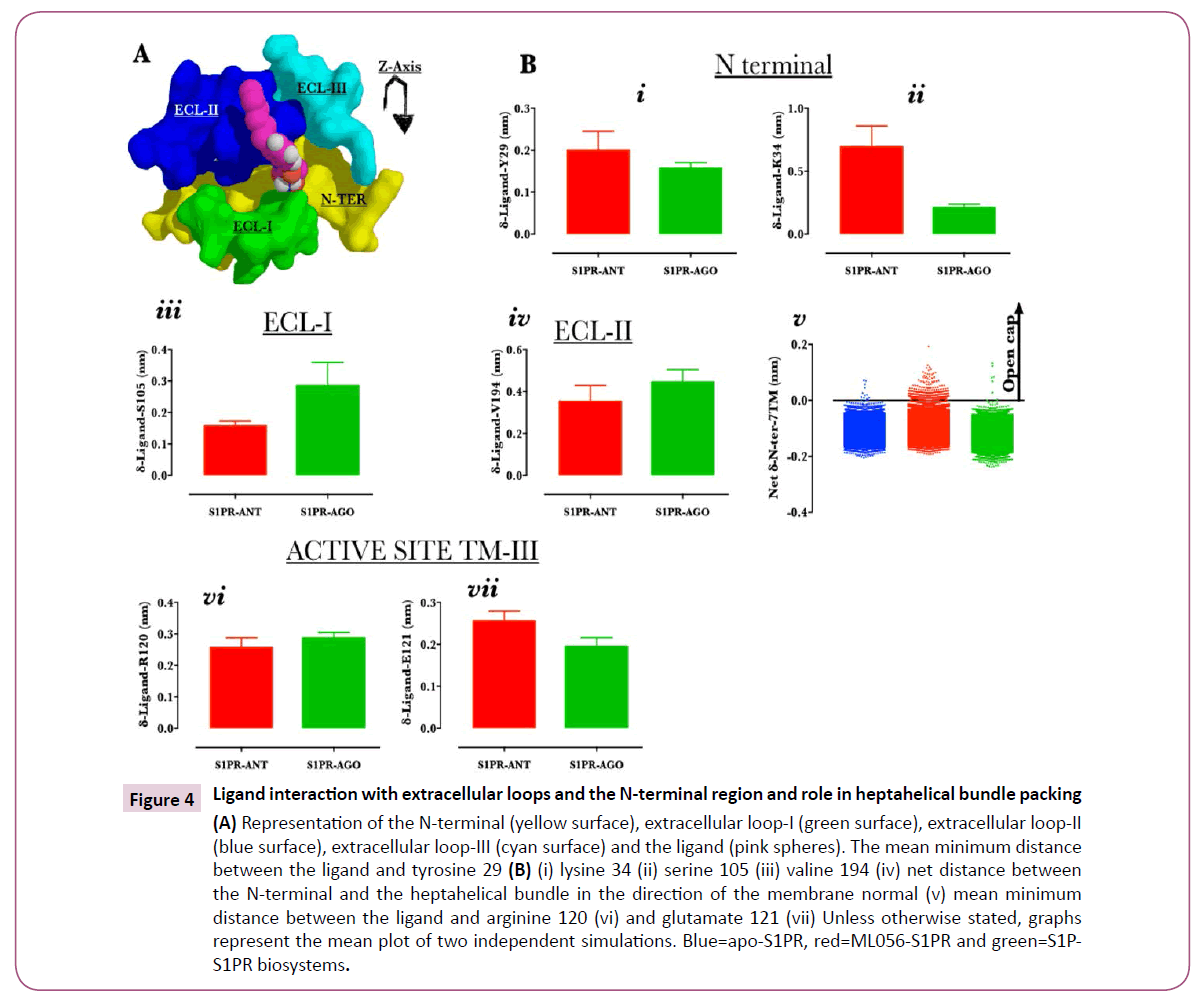
In the crystal structure (PDB ID: 3V2Y), N-terminal and the extracellular loop (ECL) (ECL-I is green, ECL-II is blue, ECL-III is cyan and N-terminal is yellow) regions observably packed closely with the TM helices (not shown) while trapping the ligand (ML056, pink sphere) (Figure 4A) into the orthosteric site [4,5]. To understand how the two ligands differentially interact with the N-terminal region, the least distance separating each of the ligands from tyrosine 29 and lysine 34 were calculated. Both ligands were located at a mean distance of 0.2 nm from tyrosine 29 (Figure 4B, i) throughout the simulation; this is consistent with the findings of Yuan et al.,[5] that both ML056 and S1P formed stable hydrogen bond interaction with tyrosine 98. Compared with ML056, sphingosine-1-phosphate showed higher residence with lysine 34 at mean distance of 0.2 nm (ML056=0.7nm, Fig. 4B, ii) as reported previously [4,5]. ECL-I residue serine 105 may dictate tight packing with ML056 and sphingosine-1-phosphate as both ligands were spaced at 0.2 and 0.3 nm from the residue (Figure 4B, iii). Similar observation was made for ECL-II as representative residue showed 0.4 nm (mean distance from valine 194, ECL-II) separation from the ligands (Figure 4B, iv). Furthermore, the net (values for structures generated along the trajectories value calculated for crystal structure, PDB ID: 3V2Y) center of mass distance of separation between the N-terminal and the heptahelical bundle (with the removal of extracellular loop residues from the calculation) was also calculated. Clearly, the data indicated that N-terminal (N1-40) was tightly packed with the heptahelical bundle in active but not in inactive S1PR (Figure 4B, v). Finally, it was observed that both ligands did not exhibit any observable difference in terms of their distance from the active site residues (arginine 120 & glutamate 121) [4] (Figure 4B, vi & vii).
Figure 4: Ligand interaction with extracellular loops and the N-terminal region and role in heptahelical bundle packing (A) Representation of the N-terminal (yellow surface), extracellular loop-I (green surface), extracellular loop-II (blue surface), extracellular loop-III (cyan surface) and the ligand (pink spheres). The mean minimum distance between the ligand and tyrosine 29 (B) (i) lysine 34 (ii) serine 105 (iii) valine 194 (iv) net distance between the N-terminal and the heptahelical bundle in the direction of the membrane normal (v) mean minimum distance between the ligand and arginine 120 (vi) and glutamate 121 (vii) Unless otherwise stated, graphs represent the mean plot of two independent simulations. Blue=apo-S1PR, red=ML056-S1PR and green=S1PS1PR biosystems.
Conclusion
The high-resolution S1PR structure resolved and deposited in protein data bank repository [4] has changed our understanding of lipid-type ligand recognition, binding, and activation of Edg family of GPCRs forever (as S1PR is the first family of this class). Following the success in crystallography, S1PR activation details were provided using microsecond molecular dynamics simulation experiments by Yuan et al [5, 20]. The major achievement of these studies was itemizing the precise contribution of key molecular events in the build up to S1PR activation, and “putting paid to” the doubts as to whether intra-helical water molecules are necessary for GPCR activation. Here, we have explored a few areas not addressed in the study such as: exploring the dihedral plasticity in aromatic residues lining the ligandbinding pocket. Interestingly, these residues displayed very distinct rotameric patterns in S1P-, ML056- and apo S1PR biosystems. Because these aromatic residues are contributed by N-terminal, extracellular loops, and the heptahelical bundle, ligand-specific and activation type rotameric signatures have been suggested. This study also identified that fully active, intermediate and inactive conformations of S1PR are accompanied by distinguishable inter-helical electrostatic interaction. While activation promoted TM1/TM4, TM2/TM7 and TM4/TM6 engagement, prior electrostatic engagements in TM3/TM4, TM3/TM6 and TM3/TM7 were dissolved. Ultimately, S1PR activation by class I ag onist followed classical GPCR activation paradigm.
Declaration of interest
This work was supported by Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan. The authors declare no conflict of interest.
References
- Rosen H, Gonzalez-Cabrera PJ, Sanna MG, Brown S (2009) Sphingosine 1-phosphate receptor signaling. Annual review of biochemistry 78: 743-768.
- Garris CS, Wu L, Acharya S, Arac A, Blaho VA, et al.(2013) Defective sphingosine 1-phosphate receptor 1 (S1P1) phosphorylation exacerbates TH17-mediated autoimmune neuroinflammation. Nature immunology 14: 1166-1172.
- Brinkmann V (2009) FTY720 (fingolimod) in Multiple Sclerosis: therapeutic effects in the immune and the central nervous system. British journal of pharmacology 158: 1173-1182.
- Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL (2012) Crystal structure of a lipid G protein-coupled receptor. Science 335: 851-855.
- Yuan S, Wu R, Latek D, Trzaskowski B, Filipek S (2013) Lipid receptor S1P(1) activation scheme concluded from microsecond all-atom molecular dynamics simulations. PLOS computational biology 9: e1003261.
- Hurst DP, Schmeisser M, Reggio PH (2013) Endogenous lipid activated G protein-coupled receptors: emerging structural features from crystallography and molecular dynamics simulations. Chemistry and physics of lipids 169: 46-56.
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic acids research 37: W623-633.
- Heifets A, Lilien RH (2010) LigAlign: flexible ligand-based active site alignment and analysis. Journal of molecular graphics &modelling 29: 93-101.
- Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL (2012) OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res 40: D370-376.
- Miller BT, Singh RP, Klauda JB, Hodoscek M, Brooks BR, Woodcock HL3rd (2008)CHARMMing: a new, flexible web portal for CHARMM. Journal of chemical information and modeling 48: 1920-1929.
- Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, et al. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29: 845-854.
- Huang J, MacKerell AD Jr (2013) CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. Journal of computational chemistry 34: 2135-2145.
- Eslami H, Mojahedi F, Moghadasi J (2010) Molecular dynamics simulation with weak coupling to heat and material baths. The Journal of chemical physics 133: 084105.
- Darden T, Perera L, Li L, Pedersen L(1999) New tricks for modelers from the crystallography toolkit: the particle mesh Ewald algorithm and its use in nucleic acid simulations. Structure 7: R55-R60.
- Cuendet MA, Van Gunsteren WF (2007) On the calculation of velocity-dependent properties in molecular dynamics simulations using the leapfrog integration algorithm. The Journal of chemical physics 127: 184102.
- Park PS, Lodowski DT, Palczewski K (2008) Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annual review of pharmacology and toxicology 48:107-141.
- Nygaard R, Zou Y, Dror RO, Mildorf TJ, Arlow DH, et al (2013) The dynamic process of beta(2)-adrenergic receptor activation. Cell 152: 532-542.
- Chiba K, Adachi k (2012) Discovery of fingolimod, the sphingosine 1-phosphate receptor modulator and its application for the therapy of multiple sclerosis. Future medchem 4: 771-781.
- Xie XQ, Chowdhury A (2013) Advances in methods to characterize ligand-induced ionic lock and rotamer toggle molecular switch in G protein-coupled receptors. Meth Enzymol 520: 153-174.
- Yuan S, Filipek S, Palczewski K, Vogel H (2014) Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Natcommun 5: 4733.
- Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, et al.(2003) Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation, Proceedings of the National Academy of Sciences of the United States of America, 100: 2290-2295.
- Rosenbaum DM, Rasmussen SG, Kobilka BK (2009) The structure and function of G-protein-coupled receptors. Nature 459: 356-363.
- Moukhametzianov R, Warne T, Edwards PC, Serrano-Vega MJ, Leslie AG, ET AL. (2011) Two distinct conformations of helix 6 observed in antagonist-bound structures of a beta1-adrenergic receptor. Proceedings of the National Academy of Sciences of the United States of America 108: 8228-8232.
- Schneider EH, Schnell D, Strasser A, Dove S, Seifert R (2012) Impact of the DRY motif and the missing "ionic lock" on constitutive activity and G-protein coupling of the human histamine H4 receptor. The Journal of pharmacology and experimental therapeutics 333: 382-392.
- Audet M, Bouvier M (2012) Restructuring G-protein- coupled receptor activation. Cell 151: 14-23.
- Dore AS, Robertson N, Errey JC, Ng I, Hollenstein K (2011) Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthinesXAC and caffeine. Structure 19: 1283-1293.
- Windh RT, Lee MJ, Hla T, An S, Barr AJ, Manning DR (1999) Differential coupling of the sphingosine 1-phosphate receptors Edg-1, Edg-3, and H218/Edg-5 to the G(i), G(q), and G(12) families of heterotrimeric G proteins. The Journal of biological chemistry 274: 27351-27358.
- Gonzalez-Cabrera PJ, Jo E, Sanna MG, Brown S, Leaf N (2008) Full pharmacological efficacy of a novel S1P1 agonist that does not require S1P-like headgroup interactions. Molecular pharmacology 74: 1308-1318.
- Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A (2012) Action of molecular switches in GPCRs--theoretical and experimental studies.Current medicinal chemistry 19: 1090-1109
- Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, et al. (2002) Beta2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. The Journal of biological chemistry 277: 40989-40996.
- Robinson PR, Cohen GB, Zhukovsky EA, Oprian DD (1992)Constitutively active mutants of rhodopsin. Neuron 9: 719-725.
- Nikiforovich GV, Marshall GR (2006) 3D modeling of the activated states of constitutively active mutants of rhodopsin. Biochemical and biophysical research communications 345: 430-437.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences