In-Silico Assessment of Various PDB Entries of Pfldh Enzyme for their Use in SBDD
Apeksha Shrivastava, Jintender Kumar, Mymoona Akhter, Mumtaz Alam M and Shaqiquzamman M
DOI10.21767/2470-6973.100016
Apeksha Shrivastava, Jintender Kumar, Mymoona Akhter*, M Mumtaz Alam and M Shaqiquzamman
Drug Design and Medicinal Chemistry Lab, Department of Pharmaceutical Chemistry, Faculty of Pharmacy and Bioinformatics Infrastructure Facility, Jamia Hamdard, New Delhi, India
- *Corresponding Author:
- Mymoona Akhter
Drug Design and Medicinal Chemistry Lab, Department of Pharmaceutical Chemistry, Faculty of Pharmacy and Bioinformatics Infrastructure Facility, Jamia Hamdard, New Delhi-110 062
E-mail: India.mymoonaakhter@gmail.com
Received date: April 27, 2016; Accepted date: May 20, 2016; Published date: May 24, 2016
Citation: Shrivastava A, Kumar J, Akhter M, et al. In-Silico Assessment of Various PDB Entries of PfLDH Enzyme for their Use in SBDD. Chem Inform. 2016, 2:1. doi: 10.21767/2470-6973.100016
Abstract
Objective: To perform in-silico structural assessment analysis of protein database entries of Plasmodium falciparum lactate dehydrogenase (PfLDH) enzyme, an important target for designing of anti-malarial drugs.
Methods: Seven PDB ID's of the enzyme PfLDH, viz. 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U were selected and downloaded from Protein Data Bank (PDB). These were subjected to Atomic Non-Local Environment Assessment (ANOLEA) energy assessment analysis and Swiss model was used to analyse Ramachandran plot.
Results: The energy assessment analysis and analysis of Ramachandran plot were carried out successfully. ANOLEA energy assessment displayed that 1T24 has 304 amino acids, only 1 high energy molecule, total non-local energy of -2722 E/Kt units and Non local normalized Z-score of -0.81. All the other PDB ID's had either more than 6 high energy amino acids or more total non-local energy or more non local normalized energy Z-Score than 1T24. Hence, it was inferred that 1T24 might be the best PDB ID amongst all. ANOLEA energy assessment analysis was also used to analyze the high energy amino acid residues of chain A of all the seven PDB’s. The total energy of PDB ID 1T24 (1.149 E/kT units) was found to be lowest and supported the inference that PDB ID 1T24 is better than others. Finally, Ramachandran plot analysis revealed that 1T24 has the highest percentage of residues in the allowed regions and only 0.4% in disallowed region and hence forth the inference was reinforced.
Conclusion: From ANOLEA energy assessment and Ramachandran plot analysis it was concluded that out of all the PDB entries, PBD 1T24 will be the best suit for carrying out structure based drug design (SBDD) studies.
Keywords
Structure assessment; ANOLEA; PfLDH; 1T2D; 1T24; 1U4O; 1U5A; 1XIV; 2A94; 4B7U
Introduction
Malaria has been tormenting the mankind since, past 5000 years causing several million deaths per year [1]. The last 15 years have observed only 37% fall in malarial incidence rates. In year 2015, 214 million new cases of malaria and 438 thousand malarial deaths were reported worldwide. But, the biggest hurdle in control of malarial incidences in maximum countries is the rapid development and expansion of insecticide and anti-malarial drug resistance [2]. Malaria is a disease caused by a parasite known as Plasmodium. There are four species of Plasmodium which commonly affect the human health, viz.,P. falciparum, P. vivax, P. malariae and P. ovale, among these P. falciparum is responsible for maximum cases of malaria. Several inhibitors have been developed for various targets, viz., Dihydrofolate reductase, Dihydropteroate synthase, Lactate dehydrogenase, Falcipain-2 and 3 etc. Lactate dehydrogenase caught our attention as it’s a relatively new receptor and also currently a lot of research is being done on it. So, it seemed to be a valid idea to analyse the PDB IDs of this enzyme [3].
Plasmodium falciparum lactate dehydrogenase enzyme
Plasmodium falciparum LDH (PfLDH) is a 316 amino acid protein, coded by a single gene on chromosome 13, and is expressed as a 1.6-kb mRNA. The amino acid sequence predicted from genomic and cDNA sequencing indicates that essential catalytic residues, such as His195, Asp168, Arg109 and Arg171 are crucial for its activity. Asp168 and His196 act as hydrogen donors; side chain of Arg171 interacts with the carboxylate of pyruvate; side chain of Arg109 interacts with the ketone oxygen of pyruvate leading to polarization of the ketone carboxyl and hydride attack from NAD; proline is critical active site residue which defines substrate and cofactor binding sites. Asn197, Lys102 and Leu163 define the conserved active site [4].
In-silico virtual screening and docking have become a very important part for the rapid design and discovery of novel drugs. But, today in this era of big-data where there are available a replete amount of PDB ID’s in the protein data bank [5], the biggest question which the drug designing chemist encounters is “Which PDB ID to select, which one is more accurate and will lead to authentic results?” This study is an attempt to answer these questions for PfLDH enzyme, which is exploited for design and development of anti-malarial drugs. Keeping in mind the importance of PfLDH in designing drugs for malaria and the significance of PDB structures in SBDD we decided to carry out the in-silico assessment analysis of PDB entries of PfLDH.
Materials and Methods
The work comprises of analysis of structure of various PDB entries of PfLDH. The study was performed on Windows 7 operating system, installed on HCL compact desktop.
Selection of PDB ID’s
PDB (Protein Data Bank) was used as a source to obtain PDB ID’s. A total of 23 PBD entries were found for “Plasmodium falciparum lactate dehydrogenase” in protein data bank [6]. The details are given in Table 1. Seven PDB’s were selected from the available 23 entries on the basis of following criteria:
| PDB ID | Description | Species | No. of mutations | Experiment type | Bound with | Resolution |
|---|---|---|---|---|---|---|
| 1CEQ | Chloroquine binds in the cofactor binding site of PfLDH. | Pf | 1 | XRD | NA | 2.00 Å |
| 1CET | Chloroquine binds in the cofactor binding site of PfLDH | Pf | 1 | XRD | Chloroquine | 2.05 Å |
| 1I10 | Human muscle 1-LDH m chain, ternary complex with NADH and oxamate | Hs | 0 | XRD | Oxamic acid and acetate ion | 2.30 Å |
| 1I0Z | Human heart L-Lactate dehydrogenase H chain, ternary complex with NADH and Oxamate | Hs | 0 | XRD | Oxamic acid | 2.10 Å |
| 1LDG | Pf 1-LDHcomplexed with NADH and oxamate | Pf | 2 | XRD | Oxamic acid | 1.74 Å |
| 1OC4 | LDHfrom Plasmodium berghei | Pb | 0 | XRD | Oxamic acid | 2.30 Å |
| 1T2C | PfLDH complexed with NADH | Pf | 0 | XRD | NA | 2.01 Å |
| 1T2D* | PfLDH complexed with NAD+ and oxalate | Pf | 0 | XRD | Oxalate ion | 1.10 Å |
| 1T2E | PfLDH S245A, A327P mutant complexed with NADH and oxamate | Pf | 2 | XRD | Oxamic acid | 1.85 Å |
| 1T24* | PfLDH complexed with NAD+ and 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | Pf | 0 | XRD | 4-hydroxy-1,2,5-oxadiazole-3-carboxylic acid | 1.7 Å |
| 1T25 | PfLDH complexed with NADH and 3-hydroxyisoxazole-4-carboxylic acid | Pf | 0 | XRD | 3-hydroxyisoxazole-4-carboxylic acid | 1.9 Å |
| 1T26 | Pf LDH complexed with NADH and 4-hydroxy-1,2,5-thiadiazole-3-carboxylic acid | Pf | 0 | XRD | 4-hydroxy-1,2,5-thiadiazole-3-carboxylic acid | 1.8 Å |
| 1U4O* | PfLDH complexed with 2,6-naphthalenedicarboxylic acid | Pf | 0 | XRD | 2,6-dicarboxynaphthalene & (4s)-2-methyl-2,4-pentanediol | 1.7 Å |
| 1U4S | PfLDH complexed with 2,6-naphthalenedisulphonic acid | Pf | 0 | XRD | 2,6-naphthalenedisulphonic acid | 2.0 Å |
| 1U5A* | PfLDH complexed with 3,5-dihydroxy-2-naphthoic acid | Pf | 0 | XRD | 3,7-dihydroxy-2-naphthoic acid | 1.8 Å |
| 1U5C | PfLDH complexed with 3,7-dihydroxynaphthalene-2-carboxylic acid and NAD+ | Pf | 0 | XRD | 3,7-dihydroxy-2-naphthoic acid | 2.65 Å |
| 1XIV* | PfLDHcomplexed with 2-({4-chloro-[Hydroxy (Methoxy) Methyl] Cyclohexyl} Amino) Ethane-1, 1, 2-triol | Pf | 0 | XRD | 2-({4-chloro-2-[hydroxy(methoxy)methyl]cyclohexyl}amino)ethane- 1,1,2-triol | 1.7 Å |
| 2A94* | Structure of PfLDH complexed to APADH. | Pf | 0 | XRD | Acetyl pyridine adenine dinucleotide, reduced | 1.5 Å |
| 2X8L | PfLDHApo structure | Pf | 0 | XRD | NA | 1.6 Å |
| 4B7U* | Pf L-lactate dehydrogenase Complexed with Bicine. | Pf | 0 | XRD | Bicine | 1.88 Å |
| 4PLZ | Crystal structure of PfLDH mutant W107fA | Pf | 1 | XRD | Oxamic acid | 1.05 Å |
| 3ZH2 | Structure of PfLDH in complex with a DNA Aptamer. | Pf/Sc | 0 | XRD | NA | 2.1 Å |
| 2HJR |
Crystal Structure of Cryptosporidium parvum malate dehydrogenase |
Cp | 0 | XRD | NA | 2.2 Å |
Italics*: selected PDB ID’s; Bold: discarded PDB ID and reason for discarding it.
NA: Not Applicable; XRD: X-RAY Diffraction; Pf: Plasmodium falciparum; Pb: Plasmodium berghei; Hs: Homo sapiens; Cp: Cryptosporidium parvum;sc:synthetic construct
Table 1: Details of PDB ID available in protein databank, their species, number of mutations, experiment type, bound ligand information, and resolution.
1. Species must be Plasmodium falciparum
2. Number of mutations must be zero.
3. X-ray crystallography is preferred method of experiment.
4. Structures which are bounded with inhibitors are preferred over the apo structures and over those which are bound with energy molecules.
5. From a particular class of PDB, the protein bounded with most diverse and most potent ligand was picked.
6. Resolution must be less than 2 Å.
7. The selected PDB IDs are shown in italics*, the discarded PDB IDs and the reason for discarding it is shown in bold in Table 1.
From the series of Azole based compounds (1T), 1T25 was discarded as the ligand had relatively low potency in comparison to other compounds of the series and 1T26 was discarded as its ligand was a bioisoester of the ligand of 1T24 and resolution of 1T24 was less than that of 1T26, hence, 1T24 was selected out of the two.
3D Structural analysis of PDB entries
All the selected PDB’s (1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U) were subjected to ANOLEA structural assessment. The 3D structures of chain A's of 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U (Figure 1) were then analysed for structure assessment by ANOLEA [7-13].
ANOLEA
ANOLEA was used to perform energy calculations on protein chain A, for evaluation of "Non- Local Environment" (NLE) of each heavy atom in the structure. In this the energy of each pairwise interaction in the non-local environment was taken from a distance-dependent knowledge-based mean force potential that was derived from a database of 147 non-redundant protein chains with a sequence identity below 25% and solved by X-Ray crystallography with a resolution lower than 3 Å [14].
ANOLEA energy assessment of PDB structures was followed by assessment of all structures by PROCHECK via swiss model-Protein Structure and Model Assessment Tool (https://swissmodel.expasy. org/) for analyzing the Ramachandran plot and for visualizing the dihedral angles ψ against ÃÆÃÂÃâââ¬Â¢ of the amino acid residues [15,16]
Results and Discussion
PfLDH being an important target for design and discovery of antimalarial drugs was selected for carrying out in-silico structural assessment analysis. Twenty three PDB’s were found in protein data bank therefore it is important to identify and utilize the most suitable structure for SBDD studies to yield good results. All the PDBs’ were studied and seven were selected for further studied to identify the most appropriate PDB ID for SBDD studies for the enzyme PfLDH.
ANOLEA energy assessment of chain A of the seven selected PDB entries of P. falciparum lactate dehydrogenase (1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U) is presented in Table 2. The table show the total number of high energy amino acids, non-local energy (E/Kt units) and Non-Local Normalized Energy Z-score for the seven selected PDB entries. It reflects that PDB ID 1T24 has only 1 high energy amino acid out of 304 amino acids and has a total non-local energy of -2722 E/kt units with second lowest non-local normalised energy Z-Score of -0.81. The PDB ID 1U4O has lowest total non-local energy and lowest nonlocal normalised energy Z-Score of -2598 E/kt units and -0.7613 respectively, but has 13 high energy amino acids out of 304 amino acids. Henceforth, the assessment reveals that chain A of 1T24 is more stable in terms of lowest number of high energy amino acids [1/304 (0.33%)], low total non- local energy (-2722 E/ kt units) and second lowest non-local normalized energy Z-score (-0.81) as the compared to the chain A’s of other PDB’s.
| PDB ID | Total Amino Acids |
High Energy Amino Acids (Percentage) | Total non-local energy (E/Kt units) |
Non-Local Normalized Energy Z-Score |
|---|---|---|---|---|
| 1T2D | 315 | 9 (2.86) | -2815 | -0.91 |
| 1T24 | 304 | 1 (0.33) | -2722 | -0.81 |
| 1U4O | 303 | 13 (4.29) | -2598 | -0.76 |
| 1U5A | 300 | 10 (3.33) | -2661 | -0.94 |
| 1XIV | 301 | 12 (3.99) | -2611 | -0.82 |
| 2A94 | 304 | 6 (1.79) | -2695 | -0.85 |
| 4B7U | 305 | 7 (2.30) | -2728 | -0.98 |
Table 2: ANOLEA energy assessment showing high energy amino acids of chain A and the total non-local energy of chain A’s of 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U.
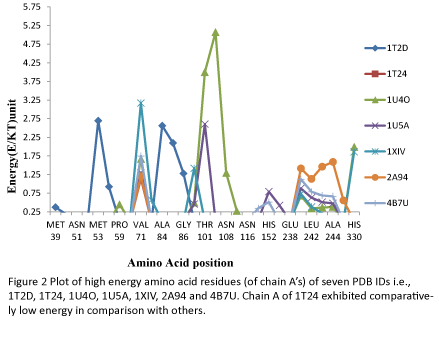
The ANOLEA energy assessment analysis was also used to analyse the high energy amino acid residues of chain A of PDB ID’s 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U, as shown in Table 3. In most of the cases the high energy amino acids were located between Ala 84 and Asn 108 followed by the amino acids located between Asn 241 and Ala 244. The other amino acids which reflected relatively higher energies were Met 53, Val 71and His 330. The total energy of PDB ID 1T24 (1.149 E/kT units) was found to be lowest, followed by 4B7U (5.881 E/kT units), 2A94 (7.362 E/kT units), 1U5A (8.118 E/kT units) and 1XIV (8.843 E/kT units). 1T2D (10.131 E/kT units) and 1U4O (17.020 E/kT units) exhibited highest total energies. Thus, the result reinforced the results obtained in Table 2 and supported the inference that PDB ID 1T24 is better than others. The energy analysis of each high energy amino acid also revealed that each of the high energy residues of 1T24 chain A has a lower energy as compared to the other chains which can also be seen in Figure 2.
| Amino acid position | 1T2D | 1T24 | 1U4O | 1U5A | 1XIV | 2A94 | 4B7U |
|---|---|---|---|---|---|---|---|
| MET 39 | 0.373 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| PRO 40 | 0.184 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| ASN 51 | 0.002 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| VPL 52 | 0.008 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| MET 53 | 2.698 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| ALA 54 | 0.925 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| PRO 59 | _ _ _ | _ _ _ | 0.452 | _ _ _ | 0.050 | _ _ _ | _ _ _ |
| ASN 70 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | 0.103 | _ _ _ | _ _ _ |
| VAL 71 | _ _ _ | 1.149 | 1.667 | 1.307 | 3.171 | 1.206 | 1.750 |
| MET 72 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | 0.567 | _ _ _ | _ _ _ |
| ALA 84 | 2.564 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| PRO 85 | 2.100 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| GLY 86 | 1.277 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| PHE 100 | _ _ _ | _ _ _ | 0.498 | 0.462 | 1.421 | _ _ _ | _ _ _ |
| THR 101 | _ _ _ | _ _ _ | 3.997 | 2.605 | _ _ _ | _ _ _ | _ _ _ |
| LYS 102 | _ _ _ | _ _ _ | 5.071 | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| ASN 108 | _ _ _ | _ _ _ | 1.292 | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| ARG 109 | _ _ _ | _ _ _ | 0.272 | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| ASN 116 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | 0.146 | _ _ _ | _ _ _ |
| GLN 151 | _ _ _ | _ _ _ | _ _ _ | 0.032 | _ _ _ | _ _ _ | 0.347 |
| HIS 152 | _ _ _ | _ _ _ | _ _ _ | 0.792 | 0.097 | _ _ _ | 0.514 |
| SER 153 | _ _ _ | _ _ _ | _ _ _ | 0.424 | _ _ _ | _ _ _ | _ _ _ |
| GLU 238 | _ _ _ | _ _ _ | 0.008 | _ _ _ | _ _ _ | _ _ _ | _ _ _ |
| ASN 241 | _ _ _ | _ _ _ | 0.684 | 0.879 | 0.733 | 1.414 | 1.127 |
| LEU 242 | _ _ _ | _ _ _ | 0.340 | 0.631 | 0.387 | 1.133 | 0.788 |
| HIS 243 | _ _ _ | _ _ _ | 0.366 | 0.509 | 0.209 | 1.465 | 0.690 |
| ALA244 | _ _ _ | _ _ _ | 0.381 | 0.477 | 0.097 | 1.589 | 0.665 |
| SER 245 | _ _ _ | _ _ _ | _ _ _ | _ _ _ | _ _ _ | 0.555 | _ _ _ |
| HIS 330 | _ _ _ | _ _ _ | 1.992 | _ _ _ | 1.862 | _ _ _ | _ _ _ |
| Total | 10.131 | 1.149 | 17.020 | 8.118 | 8.843 | 7.362 | 5.881 |
Table 3: ANOLEA energy assessment of PDB ID’s 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U reflecting high energy amino acids and their total energy.
Ramachandran plot analysis
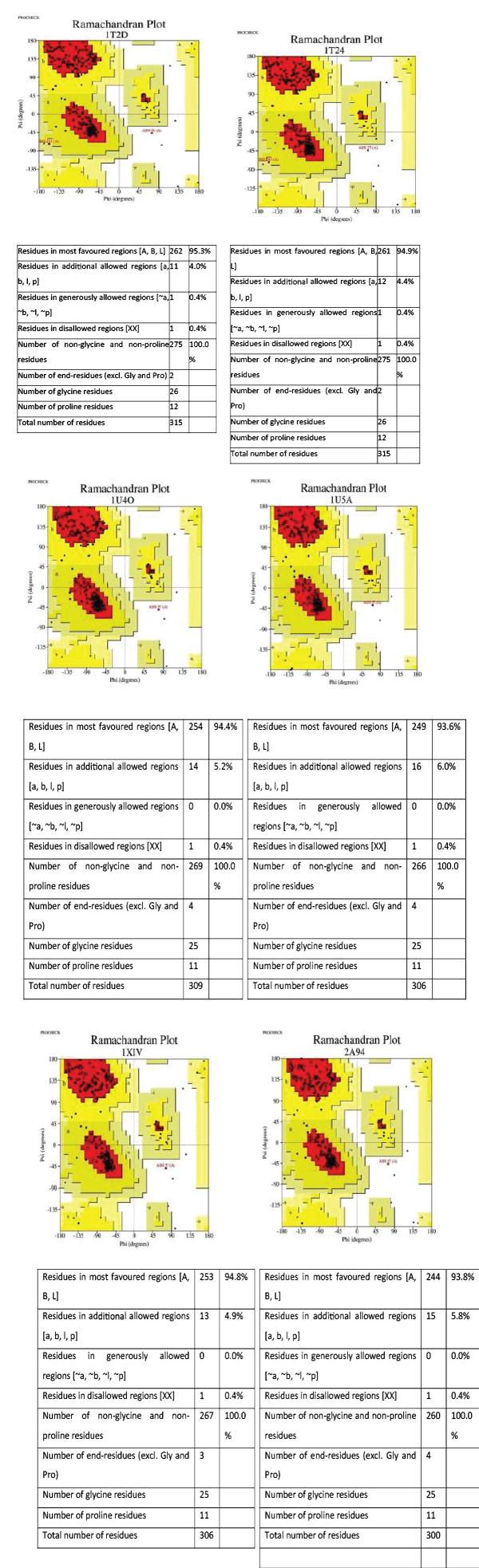
Ramachandran plot analysis (Table 4) revealed that 95.3%, 94.9%, 94.4%, 93.6%, 94.8%, 93.8%, 93.1% amino acid residues of chain A of 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U respectively were in the most favoured region of Ramachandran plot. While 4.0%, 4.4%, 5.2%, 6.0%, 4.9%, 5.8%, 6.5% of chain A of 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94, 4B7U were in the allowed regions. The plot also revealed that only one residue of chain A of 1T2D and 1T24 is in generously allowed region and one residue of chain A of 1T2D, 1T24, 1U4O, 1U5A, 1XIV and 2A94 is in the disallowed region. Ramachandran plot and its details are shown in Figure 3. It can be concluded from Ramachandran plot analysis that as 1T24 has the highest percentage of residues in the allowed regions and only 0.4% in disallowed region, it can be regarded as best amongst the PDB entries for enzyme Pf. lactate dehydrogenase.
| PDB ID | Residues in most Favoured region [A,B,L] (%) |
Residues in Additional allowed regions [a,b,l,p] (%) |
Residues in generously allowed regions [~a,~b,~l,~p] (%) |
Residues in disallowedregion (%) |
|---|---|---|---|---|
| 1T2D | 262 (95.3) | 11 (4.0) | 1 (0.4) | 1 (0.4) |
| 1T24 | 261 (94.9) | 12 (4.4) | 1 (0.4) | 1 (0.4) |
| 1U4O | 254 (94.4) | 14 (5.2) | 0 (0.0) | 1 (0.4) |
| 1U5A | 249 (93.6) | 16 (6.0) | 0 (0.0) | 1 (0.4) |
| 1XIV | 253 (94.8) | 13 (4.9) | 0 (0.0) | 1 (0.4) |
| 2A94 | 244 (93.8) | 15 (5.8) | 0 (0.0) | 1 (0.4) |
| 4B7U | 257 (93.1) | 18 (6.5) | 0 (0.0) | 0 (0.4) |
Table 4: Ramachandran plot result showing chain A’s of 1T2D, 1T24, 1U4O, 1U5A, 1XIV, 2A94 and 4B7U in favoured region, additional region, generously allowed region and disallowed region.
Conclusion
The in silico assessment analysis of PDB entries of PfLDH revealed that PDB ID 1T24 will be best to carry out SBDD studies amongst all the other entries in the Protein Data Bank, as it had the lowest energy and henceforth maximum stability. Therefore it can be utilized to produce more authenticated results for performing virtual screening and docking studies for designing of antimalarial drugs.
References
- Breman JG, Egan A, Keusch GT (2001) The intolerable burden of malaria: a new look at the numbers. Am J Trop Med Hyg 64: 4-7.
- World Health Organization (2015) World Malaria Report.WHO Global Malaria Programme. Switzerland,pp: 1-280.
- Aguiar AC, Rocha EM, Souza NB, França TC, Krettli AU (2012) New approaches in antimalarial drug discovery and development: a review. Mem Inst Oswaldo Cruz 107: 831-845.
- Richard SB, Dewar V, Clarke AR, Holbrook JJ (1997) A Model of Plasmodium falciparum lactate dehydrogenase and its implications for the design of improved antimalarials and the enhanced detection of parasitaemia. Protein Eng 10: 301-306.
- Protein Data Bank (2015) PDB Current Holdings Breakdown. National Science Foundation, USA.
- Protein Data Bank (2015) Plasmodium Falciparum Lactate Dehydrogenase Apo Structure. National Science Foundation, USA.
- Heal JW, Jimenez-Roldan JE, Wells SA, Freedman RB, Romer RA (2012) Inhibition of HIV-1 protease: the rigidity perspective. Bioinformatics 28: 350-357.
- Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, et al. (2007) Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins 67: 232-242.
- Sayer JM, Liu F, Ishima R, Weber IT, Louis JM (2008) Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J Biol Chem 283: 13459-13470.
- Adachi M, Ohhara T, Kurihara K, Tamada T, Honjo E, et al. (2009) Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography. Proc Natl Acad Sci USA 106: 4641-4646.
- Robbins AH, Coman RM, Bracho-Sanchez E, Fernandez MA, Gilliland CT, et al. (2010) Structure of the unbound form of HIV-1 subtype A protease: comparison with unbound forms of proteases from other HIV subtypes. Acta Crystallogr D Biol Crystallogr 66: 233-242.
- Reiling KK, Endres NF, Dauber DS, Craik CS, Stroud RM (2002) Anisotropic dynamics of the JE-2147-HIV protease complex: drug resistance and thermodynamic binding mode examined in a 1.09 A structure. Biochemistry 41: 4582-4594.
- Torbeev VY, Raghuraman H, Hamelberg D, Tonelli M, Westler WM, et al. (2011) Protein conformational dynamics in the mechanism of HIV-1 protease catalysis. Proc Natl Acad Sci USA 108: 20982-20987.
- Melo F, Feytmans E (1998) Assessing protein structures with a non-local atomic interaction energy. J Mol Biol 277: 1141-1152.
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures.J Appl Crystal 26: 283-291.
- Ramachandran GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. J Mol Biol 7: 95-99.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences