In-Silico Analysis of Flavin Mononucleotide Binding Domain of Nitroreductase from Pseudomonas aeruginosa PAO1
Mithil Mahale and Razia Kutty
Published Date: 2018-08-27Mithil Mahale and Razia Kutty*
Department of Microbiology, Savitribai Phule Pune University, Pune, India
- Corresponding Author:
- Razia Kutty
Department of Microbiology, Savitribai Phule Pune University, Pune 411 007, India
Tel: 305-799-753
Email: razkutty@yahoo.co.in
Received date: August 17, 2018; Accepted date: August 21, 2018; Published date: August 27, 2018
Citation: Mahale M, Kutty R (2018) In-Silico Analysis of Flavin Mononucleotide Binding Domain of Nitroreductase from Pseudomonas aeruginosa PAO1. Chem Inform Vol. 4 No. 2:2.
Copyright: © 2018 Mahale M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Objective: To carry out in-silico sequence and structural analysis of flavin mononucleotide binding domain (FMN) of Pseudomonas aeruginosa nitroreductase enzyme which is of biotechnological significance and Comparison with its structural analogues
Methods: The FMN domain from Pseudomonas aeruginosa nitro reductase gene was cloned and sequenced. The sequence was subjected to multiple alignment, homology modeling and Ramchandran analysis with close structural homologues of nitro reductase as well as with that of phylogenetically related species.
Results: FMN binding site in case of Pseudomonas aeruginosa nitro reductase is more like the structural analogues B. subtilis, T. thermophilus and S. pneumoniae nitro reductases; conservation scores are mostly>6. However, the dimer interface sites are observed to be variable in nature. The phylogenetically related and diverse FMN domains were also analyzed with respect to conservation and variation of sequence
Conclusions: The conservation or variations of amino acids at the dimer interface would have implications in varied conformations of FMN binding domain inferring difference in function and kinetics of catalysis.
Keywords
Nitro reductase; Flavoproteins; Flavin mononucleotide; Homology modelling
Introduction
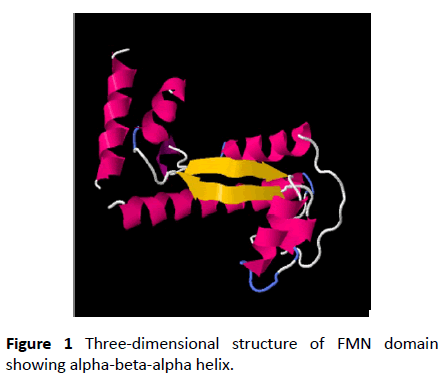
Nitro reductase enzyme are flavoproteins found in all kingdoms. They catalyze the reduction of nitro compounds using NADPH as source of reducing power and FMN as redox co-factor [1-4]. Nitro reductase enzymes possesses broad substrate specificity and follows ping-pong mechanism. The isoalloxazine ring in FMN can accept and donate one or two electrons in biological reactions. The conserved domain analysis shows present of a single domain which has the FMN binding domain, NADPH binding domain and interface region where the homodimeric unit interacts [5-8]. The FMN cofactor incorporated in flavoproteins as a posttranslational modification it might be of significance in oxidoreductase functions. To study the molecular mechanism involved in flavoproteins, site directed mutagenesis is adopted as a favored technique which minimally disrupts the interaction between FMN, NADPH and the apoprotein. However, mutation of the same amino acids in similar enzymes do not cause the same functional changes. There is no universal rule which can be accepted for mutation studies. Substitution of residues cannot be based only on chemical properties the confirmation and the folding requirements play a major role [9]. Hence comparison of domains of structures of reported proteins with respect to confirmation or flexibility will give insights to structure function correlation of flavoproteins (Figure 1).
This study attempts to address the significance of the variations observed in the FMN binding and dimer interface sites in case of Pseudomonas aeruginosa PAO1 nitro reductase.
Materials and Methods
The work done is comprised of sequence analysis, multiple sequence alignment, 3-D structure using homology modeling and characterization of secondary structures of the proteins of closely related species along with their Ramchandran analysis to identify the protein folds.
Construction of phylogenetic tree and selection of clade
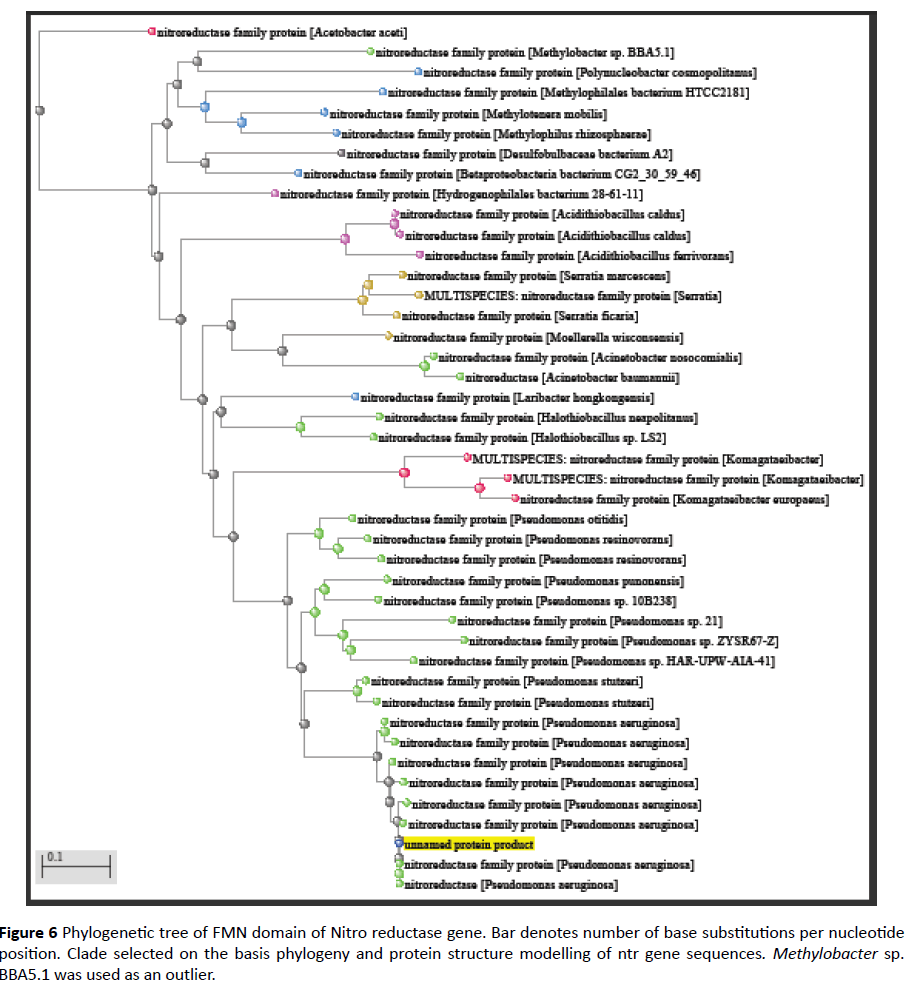
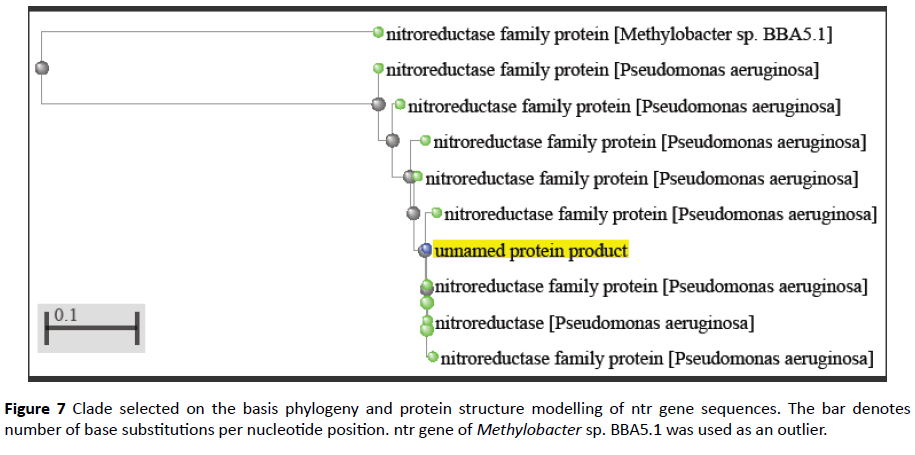
The protein query sequence of FMN domain was subjected to BLAST analysis using NCBI-PSI BLAST [10]. Phylogenetic tree was constructed based on the sequence similarity up to 40%. Later, a clade was selected from this tree for further analysis. Methylobacter spp. BBA5.1 was taken as an outlier. Multiple Sequence Alignment was done of those protein sequences.
Homology modeling and secondary structure prediction
The sequences obtained by MSA were then used for determining the 3-D structure of the proteins in those clades. This was done using Protein Homology/Analogy Recognition Engine 2 (PHYRE 2) software [11]. The individual sequences were also converted into PDB format to view their 3-D structure using RasMol or CHIMERA. PHYRE 2 software also provided the information regarding homology modeling between the protein structures of the organisms under study. The secondary structure determination was also performed using this software that clarified the region of conserved amino acid residues in the individual protein.
Ramchandran analysis
Ramchandran plot was constructed of the proteins under study by putting the amino acid sequence in the RAMPAGE software that gave an idea about the torsional folds and the conformation between the angles of amino acids in those proteins [12].
Results and Discussion
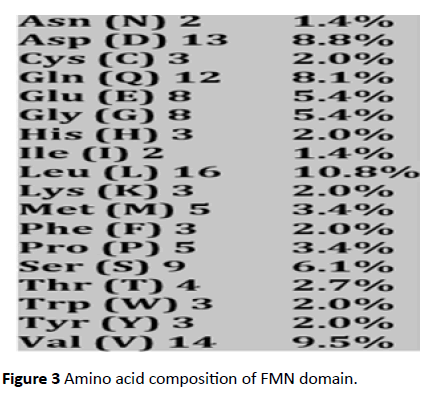
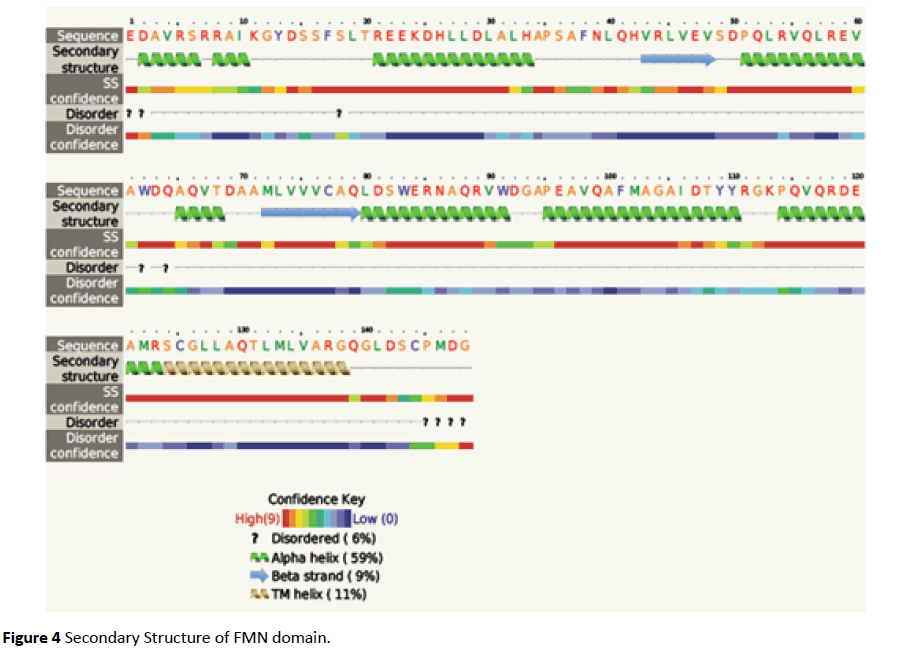
The FMN binding domain from nitro reductase gene was amplified by PCR and cloned in PUC18 vector and further sequenced (Figures 2 and 3). Secondary structure analysis by PHYRE2 shows alpha helix (59%) and Beta strand (9%) (Figure 4). The 3D structure based on homology modelling shows the characteristic alpha beta alpha structure of nitro reductase family (Figure 4).
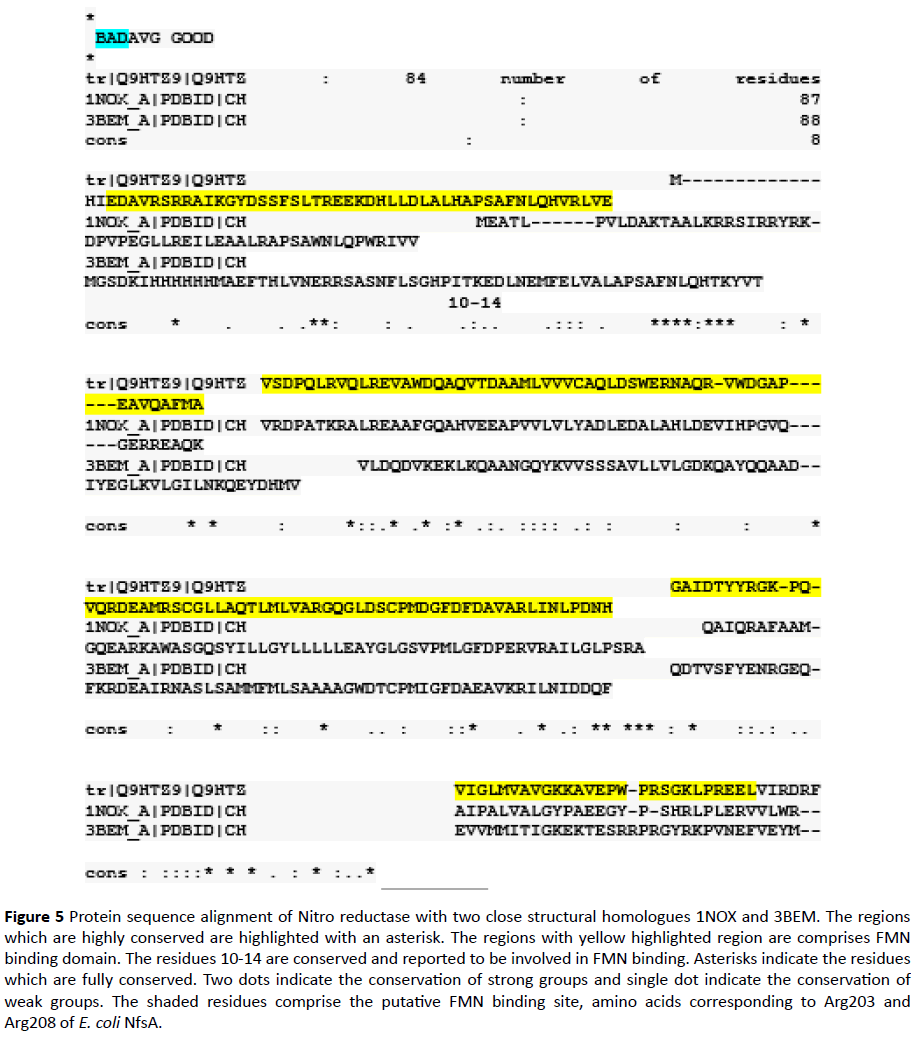
In this study an In-silico analysis of the FMN binding domain of nitro reductase enzyme from Pseudomonas aeruginosa and its close structural analogues (obtained from database) has been done to understand structure function correlation with well-studied homologue NADH Oxidase (PDB ID 1NOX) (Table 1) (Figure 5). Comparison of residues from the four homologues in the present study indicated that the FMN binding site in case of PaNR is more similar as observed in case of B. subtilis, T. thermophilus and S. pneumoniae nitro reductase; conservation scores are mostly>6 (Table 1). However, the dimer interface sites are observed to be variable in nature (Table 2). As previously studied the stretch of residues binding to FMN changes its conformation in both reduced and oxidized states which is responsible for electron transfer and rebinding of FMN to the apoprotein [13-17]. Thus, the variation in residues could have implications in binding of FMN to the monomers and changes in enzyme activity. FMN plays diverse role, its effect on antioxidant activity in Deinococcus radiodurans under H2O2 stress has been documented [18,19]. This residue can be categorized into different types of motifs. These variations are correlated with modeled structures of FMN domain in terms of Ramchandran analysis parameters. The phylogenetically close FMN domain can show significant variation in allowed and disallowed residues (Table 3). Analysis of residues present in disallowed main chain confirmation or multiple conformations can be investigated further for the redox characteristics of proteins.
Figure 5 Protein sequence alignment of Nitro reductase with two close structural homologues 1NOX and 3BEM. The regions which are highly conserved are highlighted with an asterisk. The regions with yellow highlighted region are comprises FMN binding domain. The residues 10-14 are conserved and reported to be involved in FMN binding. Asterisks indicate the residues which are fully conserved. Two dots indicate the conservation of strong groups and single dot indicate the conservation of weak groups. The shaded residues comprise the putative FMN binding site, amino acids corresponding to Arg203 and Arg208 of E. coli NfsA.
| PDB ID | Source | %Query Coverage | %Similarity | %Identity | Resolution in Angstrom |
|---|---|---|---|---|---|
| 3BEM | B. subtilis | 94 | 50 | 29 | 1.65 |
| 1NOX | Thermus thermophilus | 96 | 49 | 34 | 1.59 |
| 2B67 | Streptococcus pneumoniae | 82 | 45 | 25 | 2.05 |
| 1ICV | E. coli | 92 | 43 | 25 | 2.4 |
Table 1: Comparison of PaNR sequence with four structural homologs (Single column fitting Table).
| Site | Residue position as per 3BEM | 3BEM | 1NOX | 2B67 | 1ICV | Pseudo_NR | Conservation score | Other variable residues |
|---|---|---|---|---|---|---|---|---|
| FMN binding | 11 | R | R | R | R | R | 9 | H, A, R |
| 13 | S | S | A | S | A | 9 | A, S, T, D | |
| 39 | P | P | P | P | P | 9 | S, A, P | |
| 40 | S | S | S | S | S | 9 | A, S, T, D | |
| 41 | A | A | A | S | A | 9 | H, A, S, P, G, V | |
| 43 | N | N | N | N | N | 8 | A, F, S, N, K, Y, H, M, D, G | |
| 70 | K | H | Q | K | Q | 6 | F, A, W, N, K, Y, V, H, Q, M, R, G, L | |
| 137 | L | I | L | L | L | 7 | H, A, M, C, I, L | |
| 154 | C | V | N | V | C | 8 | A, T, N, C, I, G, V | |
| 155 | P | P | I | P | P | 6 | S, F, A, W, T, P, Y, V, M, I | |
| 157 | I | L | L | E | D | 7 | A, S, T, I, G, L, E | |
| 158 | G | G | G | G | G | 8 | A, S, P, G, V | |
| 159 | F | F | F | F | F | 6 | A, F, T, K, I, Y, L, V | |
| Dimer interface | 7 | L | A | L | V | A | 5 | S, A, T, E, V, M, D, I, L |
| 8 | V | A | N | A | V | 5 | A, F, N, Y, V, M, I, L | |
| 11 | R | R | R | R | R | 9 | H, A, R | |
| 29 | N | R | R | E | D | 1 | S, A, T, N, K, E, H, Q, M, D, R | |
| 36 | A | L | T | Q | L | 6 | A, T, N, K, V, Q, M, D, R, I, G, L | |
| 37 | L | R | L | Y | H | 3 | A, S, F, T, W, N, K, Y, Q, M, C, R, L | |
| 38 | A | A | A | S | A | 9 | S, A, T, G, V | |
| 43 | N | N | N | N | N | 8 | A, F, S, N, K, Y, H, M, D, G | |
| 45 | Q | Q | Q | Q | Q | 8 | S, T, Y, E, H, Q, C, R | |
| 49 | Y | I | F | F | L | 7 | F, I, Y, V | |
| 50 | V | V | V | I | V | 7 | F, T, C, I, Y, L, V | |
| 51 | T | V | V | V | E | 7 | F, W, T, N, Y, V, R, I, G, L | |
| 53 | L | R | R | S | S | 5 | S, T, N, K, E, H, Q, M, D, R, L | |
| 54 | D | D | E | T | D | 7 | S, T, D, N, E | |
| 55 | Q | P | K | E | P | 2 | S, A, N, K, P, E, V, Q, M, D, R, G | |
| 136 | S | Y | G | Y | G | 7 | S, A, T, P, G, Y | |
| 137 | L | I | L | L | L | 7 | H, A, M, C, I, L | |
| 144 | L | L | L | L | L | 6 | A, H, M, T, I, L, V | |
| 145 | S | L | A | S | V | 8 | S, A, M, T, G, L | |
| 148 | A | A | D | A | G | 8 | S, A, T, N, V, Q, C, D, R, G, L | |
| 168 | L | L | L | F | I | 8 | F, N, G, L, V | |
| 169 | N | G | E | G | N | 1 | S, F, A, N, K, E, V, Q, D, R, I, G, L | |
| 195 | Y | H | Y | S | G | 6 | S, T, N, K, P, E, Y, V, H, D, R, L | |
| 196 | R | R | R | R | K | 9 | A, N, R, E, V | |
| 197 | K | L | L | L | L | 5 | S, P, K, Y, E, Q, D, I, R, L | |
| 199 | V | L | V | Q | E | 1 | A, F, K, E, Y, V, H, Q, I, G, L | |
| 200 | N | E | D | N | D | 4 | A, S, N, K, E, H, Q, D | |
| 201 | E | R | E | I | E | 3 | A, T, K, E, V, H, Q, M, D, R, I, G, L | |
| 202 | F | V | I | T | N | 7 | F, A, W, I, L, V |
Table 2: The residues in the FMN binding site and dimer interface among the four Nitroreductase sequence (PaNR, 3BEM, 1NOX, 2B67 and 1ICV) along with the conservation is listed. The sites conserved among all the four sequences are highlighted in red while semi-conserved sites are shown in blue. (2 Column fitting Colored Table).
| Sr. no | Query Sequences | Ramchandran analysis | ||
|---|---|---|---|---|
| No. of residues in favoured region (~98% expected) |
No. of residues in allowed region (~2% expected) |
No. of residues in Outlier region | ||
| 1 | FMN domain | 132 (96.4%) | 5 (3.6%) | 0 |
| 2 | P1 (>WP_074207593) |
131 (90.3%) | 13 (9.0%) | 1 (0.7%) |
| 3 | P2 (>WP_034026955) |
132 (91%) | 12 (8.3%) | 1 (0.7%) |
| 4 | P3 (>WP_024917716) |
132 (91%) | 12 (8.3%) | 1 (0.7%) |
| 5 | P4 (>WP_058178814) |
133 (91.7%) | 10 (6.9%) | 2 (1.4%) |
| 6 | P5 (>WP_003153367) |
131(90.3%) | 13 (9.0%) | 1 (0.7%) |
| 7 | P6 (>WP_043158434) |
127 (90.7%) | 11 (7.9%) | 2 (1.4%) |
| 8 | P7 (>WP_058162654) |
135 (93.1%) | 9 (6.2%) | 1 (0.7%) |
| 9 | P8 (>WP_086340576) |
129 (94.2%) | 6 (4.4%) | 2 (1.5%) |
| 10 | Methylobacter sp. BBA5.1 (>WP_051961828) |
136 (93.8%) | 7 (4.8%) | 2 (1.4%) |
Table 3 Ramchandran analysis of FMN domain of organisms containing Ntr gene. Note: P1 refers to first species of Pseudomonas aeruginosa from the clade under study and likewise. The outlier selected was Methylobacter sp. BBA5.1 (>WP_051961828)
Comparative analysis is also done from phylogenetically close and distant species in order to gain information of evolutionary patterns of FMN domain as documented earlier [20] (Figures 6 and 7). This analysis demonstrates close relation with Pseudomonas clade followed by multispecies and Methylobacter.
Flavoproteins are the proteins containing flavin moiety viz. FMN or FAD as the redox co-factor which functions as electron carrier during enzymatic reaction. Nitro reductases, CytochromeP450 monooxygenases, amine oxidase are flavoproteins of relevance in biological reactions. The energy producing reactions are dependent on flavins as they can convert two electrons process to one electron reduction. The oxidoreduction reaction is mediated by the interconversion of the isoalloxazine moiety of co-factor in oxidized and reduced state. Due to this redox property flavins are involved in many biochemical reactions. The flavins are mostly bound to the apoproteins by non-covalent bonds. There are few flavoproteins with flavins bound covalently. The studies reporting the mechanism of flavoproteins have shown that the FMN cofactor undergoes changes in protonation state and affects the conformation of FMN binding domain. Whereas the affinity of flavoproteins to the flavins depends on the amino acid residues as demonstrated by mutation studies.
The conservation or variations of amino acids would be reflected in varied conformational changes of FMN binding domain and hence different binding affinity among enzymes [19]. The experimental validation of the conformational changes and redox potential will imply the significance of this variations observed.
Conclusions
The FMN domain of nitro reductase from PAO1 was cloned and sequenced. The conservation of amino acid residues with structural analogues of nitro reductase and variations at dimer interface can be explored for functional differences among structural analogues. The 3D structure of the domain shows characteristic alpha beta alpha helix like nitro reductase. This information would have implications in varied redox potentials of FMN binding domain.
References
- RCSB PDB (2002) Structure of Nitroreductase from E. cloacae Bound with 2e-Reduced Flavin Mononucleotide (FMN). Available from: https://www.rcsb.org/structure/1KQD.
- RCSB PDB (2017) Structure of nitroreductase from E. cloacae complexed with para-nitrobenzoic acid. Available from: https://www.rcsb.org/pdb/explore/litView.do?structureId=5J8G.
- RCSB PDB (2000) Nitroreductase from Enterobacter cloacae. Available from: https://www.rcsb.org/structure/1nec.
- Verma R, Schwaneberg U, Roccatano D (2012) Conformational Dynamics of the FMN-Binding Reductase Domain of Monooxygenase P450BM-3. Journal of Chemical Theory and Computation 9: 96-105.
- Isayev O, Crespo-Hernández C, Gorb L, Hill F, Leszczynski J, et al. (2012) In silico structure-function analysis of E. cloacae nitroreductase. Proteins: Structure, Function and Bioinformatics 80: 2728-2741.
- Joosten V, Van Berkel W (2007) Flavoenzymes. Current Opinion in Chemical Biology 11: 195-202.
- Nivinskas H, Koder R, Anusevičius Ž, Šarlauskas J, Miller A, et al. (2001) Quantitative Structure–Activity Relationships in Two-Electron Reduction of Nitroaromatic Compounds by Enterobacter cloacae NAD(P) H: Nitroreductase. Archives of Biochemistry and Biophysics 385: 170-178.
- Roldán M, Pérez-Reinado E, Castillo F, Moreno-Vivián C (2008) Reduction of polynitroaromatic compounds: The bacterial nitroreductases. FEMS Microbiology Reviews 32: 474-500.
- Hefti M, Vervoort J, Van Berkel W (2003) Deflavination and reconstitution of flavoproteins. European Journal of Biochemistry 270: 4227-4242.
- Altschul S (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389-3402.
- Zhao Q, Modi S, Smith G, Paine Z, Mcdonagh P, et al. (1999) Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93a resolution. Protein Science 8: 298-306.
- Ramachandran G, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology 7: 95-99.
- Verma R, Schwaneberg U, Roccatano D (2012) Conformational Dynamics of the FMN-Binding Reductase Domain of Monooxygenase P450BM-3. Journal of Chemical Theory and Computation 9: 96-105.
- Weikl T, Paul F (2014) Conformational selection in protein binding and function. Protein Science 23: 1508-1518.
- Yang P, Chen Z, Shan Z, Ding X, Liu L, et al. (2014) Effects of FMN riboswitch on antioxidant activity in Deinococcus radiodurans under H2O2 stress. Microbiological Research 169: 411-416.
- Krogh A, Larsson B, Von Heijne G, Sonnhammer E (2001) Predicting transmembrane protein topology with a hidden markov model: application to complete genomes by F. Cohen. Journal of Molecular Biology 305: 567-580.
- Driggers C, Dayal P, Ellis H, Karplus P (2014) Crystal Structure of Escherichia coli SsuE: Defining a General Catalytic Cycle for FMN Reductases of the Flavodoxin-like Superfamily. Biochemistry 53: 3509-3519.
- Ellis H (2010) The FMN-dependent two-component monooxygenase systems. Archives Biochem Biophy 49: 1-12.
- Seo M, Park J, Kim E, Hohng S, Kim H (2014) Protein conformational dynamics dictate the binding affinity for a ligand. Nature Communications, p. 5.
- Paine M, Garner A, Powell D, Sibbald J, Sales M, et al. (2000) Cloning and Characterization of a Novel Human Dual Flavin Reductase. Journal of Biological Chemistry 275: 1471-1478.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences