Identification of novel drug targets and lead compounds in Anthrax and Pneumonia causing pathogens using an In Silico approach
Varun Khanna, Ashutosh Kumar, Rishi Shanker
Varun Khanna1*, Ashutosh Kumar1, Rishi Shanker1
Institute of Life Sciences, Ahmedabad University, Navrangpura, University Road, Ahmedabad, Gujarat, India
- *Corresponding Author:
- Varun Khanna
Institute of Life Sciences, Ahmedabad University
Navrangpura, University Road
Ahmedabad, Gujarat
India
Tel: +91 79 26302414-18
Fax: +91 79 26302419
E-mail:varun.khanna@ahduni.edu.in
Received date: June 30, 2015; Accepted date: August 19, 2015; Published date: August 24, 2015
Citation: Khanna , Kumar A, Shanker R. Identification of novel drug targets and lead compounds in Anthrax and Pneumonia causing pathogens using an In Silico approach. Chem Inform. 2015, 1:1.
Abstract
Increased resistance to antibiotics and delay in treatment significantly reduce the chances of survival for anthrax and pneumonia patients. In the present study comparative proteome analysis and virtual screening approaches were used to discover novel therapeutic drug targets and antibacterial lead compounds for Bacillus anthracis and Bacillus cereus strains responsible for causing anthrax and pneumonia, respectively. The employed systematic workflow identified a total of 803 proteins, within the genomes of the aforesaid pathogens, with no significant sequence similarity to human proteins. Out of 803 non homologous proteins, 10 were found to be essential for the survival of pathogens. Further based on number of criteria including essentiality, druggability and availability of structural information thioredoxin reductase (TrxR) was shortlisted as the potential drug target. In silico docking studies with three open source docking programs identified four potential inhibitors of bacterial TrxR.. In conclusion this study resulted in identification of a novel drug target (Thioredoxin reductase) for Bacillus anthracis and Bacillus cereus previously not known of these pathogenic microorganisms) and potential inhibitors of TrxR that could facilitate in selection of drug leads for entry into drug design and production pipelines. . In addition, the methodology used can be applied for the identification of new drug targets and drug leads in case of other pathogenic microorganisms.
Keywords
Bacillus, Anthrax, Pneumonia, Drug target identification, Molecular docking, Thioredoxin reductase inhibitors
Introduction
The arrival of post-genomic era has ushered the possibility of high-throughput screening of compound libraries against all the potential targets of a pathogen. However, a persistent bottleneck in structure based drug discovery is the availability of druggable target which play a vital role in parasite and is sufficiently different from human counterpart. Experimental validation of identified potential drug targets is an additional challenge, as it is impractical for genome wide scales. Therefore, several in silico approaches such as discovering key steps in metabolic pathways, chemogenomics, subtractive and comparative genomics have been proposed to identify and prioritize drug and vaccine targets for different pathogens [1-4].
Following is a brief summary of studies on drug target identification and validation. In one of the studies a combination of in silico and experimental approaches identified 52 proteins as probable vaccine candidates for highly invasive Indian Streptococous pyogenes serotypes [5]. This study also revealed that the highly invasive group A Streptococous M49 had metabolically more active membrane associated protein machinery than less invasive M1 strain. Caffrey et al. [6] compared the putative proteome of Schistosoma mansoni with the two model organisms Caenorhabditis elegans and Drosophila melanogaster using Genlight software. The authors were able to identify 35 druggable proteins in S. mansoni, 18 of which were homologous to the proteins with 3D structures including co-crystallized ligands. The in silico and in vitro screening for potential vaccine candidates and virulence related genes in the B. anthracis virulence plasmid pXO1 has previously been investigated in [7]. The known 143 open reading frames (ORFs) in pXO1 were subjected to comparative genomic analysis with related genomes of Bacillus subtilis, Bacillus halodurans, Bacillus cereus and the pBtoxis plasmid of Bacillus thuringiensis var. Israeliensis. As a consequence, authors were able to annotate more than 60% ORFs compared to 30% known earlier. Subsequently, nine ORFs were successfully expressed as full length peptides and three were found to be antigenic with immunogenic potential [7]. Crowther et al. [8] reviewed different strategies to prioritize pathogen proteins based on the properties which are considered desirable for drug targets. The authors also suggested novel drug targets for seven tropical disease pathogens including Mycobacterium tuberculosis, Plasmodium falciparum and others.In the current study, in silico approach was used to discover novel drug targets in anthrax and pneumonia causing strains and substrains of Bacillus. The discovered targets were further subjected to virtual screening in order to identify novel lead compounds inhibiting those targets. We used three strains of Bacillus (viz. B. anthracis, B. cereus and B. toyonensis) which are Gram-positive, endospore-forming, rod-shaped bacteria and have a similar genetic background. B. anthracis (the causative agent of anthrax) exist in the environment as a spore and can remain viable in soil for decades [9]. Spores ingested by grazing herbivores germinate within the animal and produce the virulent vegetative forms that replicate and eventually kill the host. Products (e.g., meat or hides) from infected animals serve as a reservoir for human disease [9]. Recently, there has been an increase in the number of cases of “injection anthrax" in European countries, including Denmark, France, and the United Kingdom (a form of disease that affects heroin users and is caused by B. anthracis) [10]. B. cereus on the other hand is an opportunistic, non-specific human pathogen responsible for causing pneumonia [11,12], food poisoning [13] nosocomial infections and contamination in hospitals [14]. It frequently pollutes dairies due to toxin synthesis and formation of heat−resistant spores, which survive pasteurization [15]. It is also considered as one of the most severe organism that affect the eye (most often after trauma) resulting in blindness [16]. B. toyonensis is a non-pathogenic strain which was first isolated in Japan in 1966. It is being used as a probiotic in animal feed since 1975 [17].
The existing drugs and vaccines currently available for the treatment of anthrax disease and pneumonia are effective only when the treatment is initiated soon after early diagnosis. However, delay in treatment significantly reduces the chances of survival. Another problem with most of the existing antimicrobial drugs is their inability to act on the latent forms of bacteria. This strongly suggests the need for new therapeutic targets and novel drug or vaccine candidates that offer better protection.
Therefore, in the present study, we predicted and validated novel drug targets using in silico approaches. Additionally, we screened a library of compounds from ZINC database to predict lead molecules of high binding affinity and selectivity for the identified targets. The approach used, can be applied for the identification of novel drug targets and drug leads in case of other pathogenic microorganisms.
Material and methods
Data assembly, clustering and identification of orthologous proteins
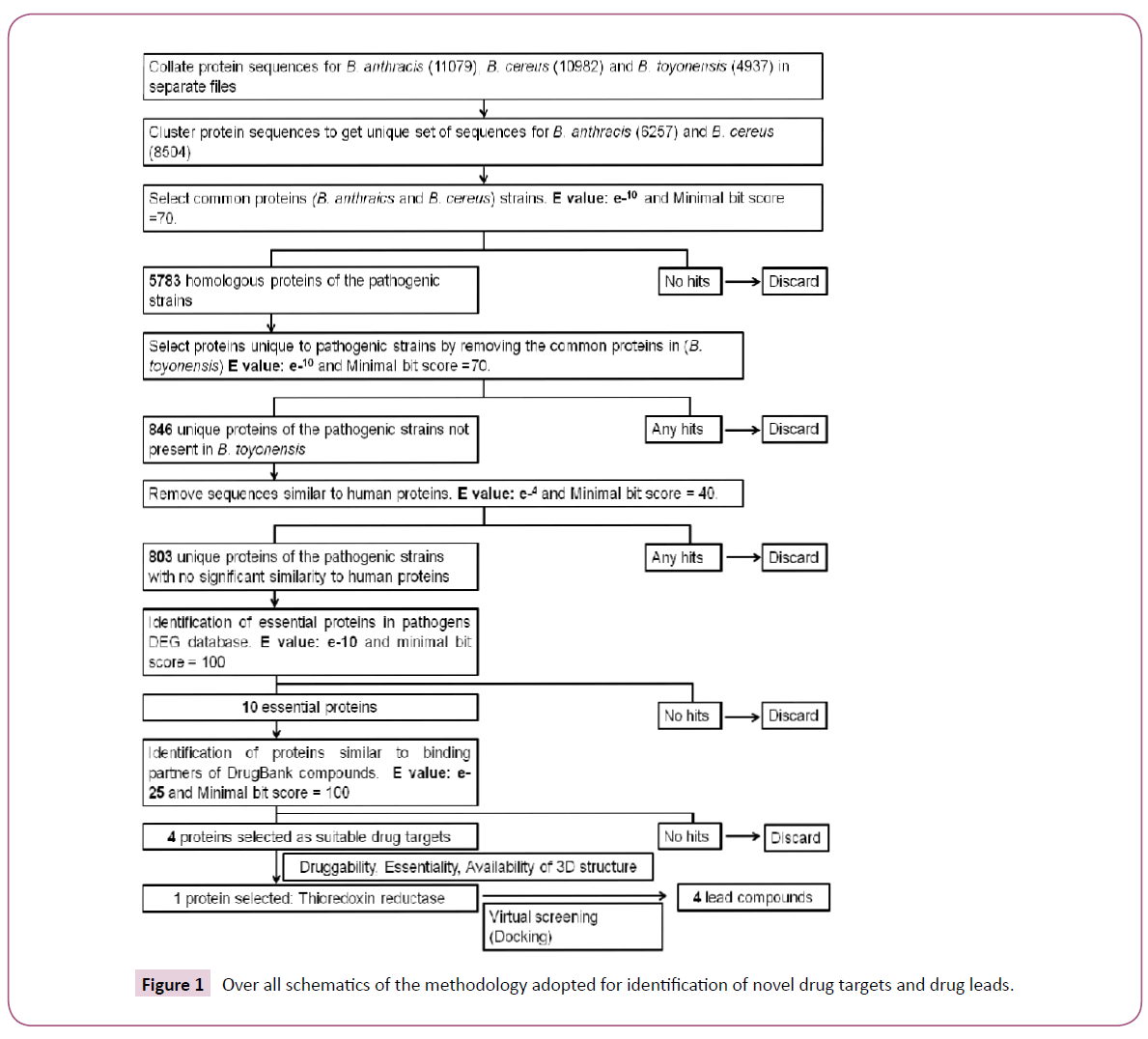
A systematic workflow involving various bioinformatics methods, tools and databases for identification of novel drug targets in pathogenic strains of B. anthracis and B. cereus was designed (Figure 1). The complete proteomes for B. anthracis (strain A0248 and H9401), B. cereus (strain B4264 and 03BB102) and B. toyonensis were downloaded from Uniprot database (www. uniprot.org). 5291 and 5788 protein sequences from strain A0248 and H9401 respectively were combined in one file. Similarly, 5598 and 5384 protein sequences from B. cereus strain 03BB102 and B4264 respectively were merged in other file. Using single linkage clustering algorithm implemented in BLASTClust tool available from NCBI-BLAST package [18], datasets were clustered using 90% sequence coverage and 95% sequence identity. The program begins with pairwise matches and places a sequence in a cluster if the sequence matches at least one member in the cluster. Clustering was performed in order to remove redundancy and get a unique set of sequences from B. anthracis and B. cereus strains. For identification of orthologs clustered sequences were then subjected to BLASTP analysis with the e-value inclusion threshold set to e-10. A subsequent BLAST search against the nonpathogenic strain (B. toyonensis) elicited the set of sequences that were present in pathogenic strains but were absent in nonpathogenic strain.
Identification of non-homologous and essential proteins
Protein sequences exclusive to pathogenic strains were compared with the non-redundant database of human proteins using BLASTP to reveal non homologous set of proteins. Sequences with no significant sequence similarity to human proteins (non homologous set) were further filtered by searching for homologous sequences in the database of essential genes (DEG) [19]. The DEG database search was performed to identify the critical genes necessary for the survival of pathogens. The hits with e-value less than e-10 and with the minimum bitscore of 100 having same name and function were considered as homologous to essential proteins. The e-value used in this study was as described by Gosh et al. [20].
Identification of potential drug targets
In order to identify potential drug targets, identified essential proteins were aligned to the targets of all FDA-approved, small molecule, nutraceutical and biotech drugs in DrugBank (DB) database [21]. The alignments with e-value less than (1E-25) were discarded except for two proteins (UniProtID: I0D343 and I0D5Y2) because of the availability of their counterparts in other pathogenic organisms present in DB database. To further narrow down our search for the potential drug targets we searched for the availability of X-ray structures of proteins bound with a natural ligand or a drug in the Protein Data Bank (PDB) [22]. Structural information about the protein target could enhance the drug discovery by facilitating the structure based drug design including docking and high throughput virtual screening.
Virtual screening
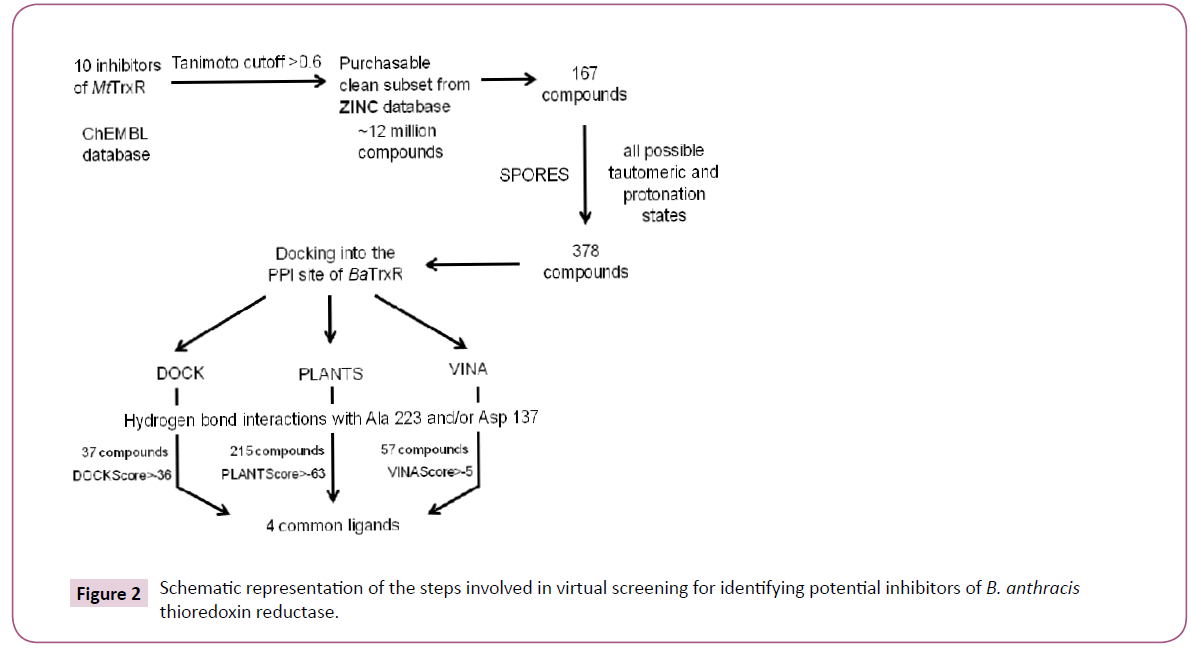
The virtual screening workflow has been described in Figure 2. We narrowed down to a single target called thioredoxin reductase (TrxR) for the reasons discussed under results and discussion section. Initially ten inhibitors of M. tuberculosis thioredoxin reductase (MtTrxR) with known IC50 values were taken from the ChEMBL database [23]. Subsequently, a library was created by exhaustively searching ~12 million compounds of the purchasable clean subset from the ZINC database. This yielded 167 compounds with the similarity score > 0.6. Similarity was measured using Tanimoto coefficient on the basis of extended connectivity fingerprints implemented in SciTegic Pipeline Pilot. Finally, virtual screening campaign was launched to identify potential small molecule inhibitors of B. anthracis thioredoxin reductase (BaTrxR) that target the protein-protein interaction site (PPI) for the substrate thioredoxin (Trx).
Structural comparison of Bacillus anthracis TrxR (BaTrxR)
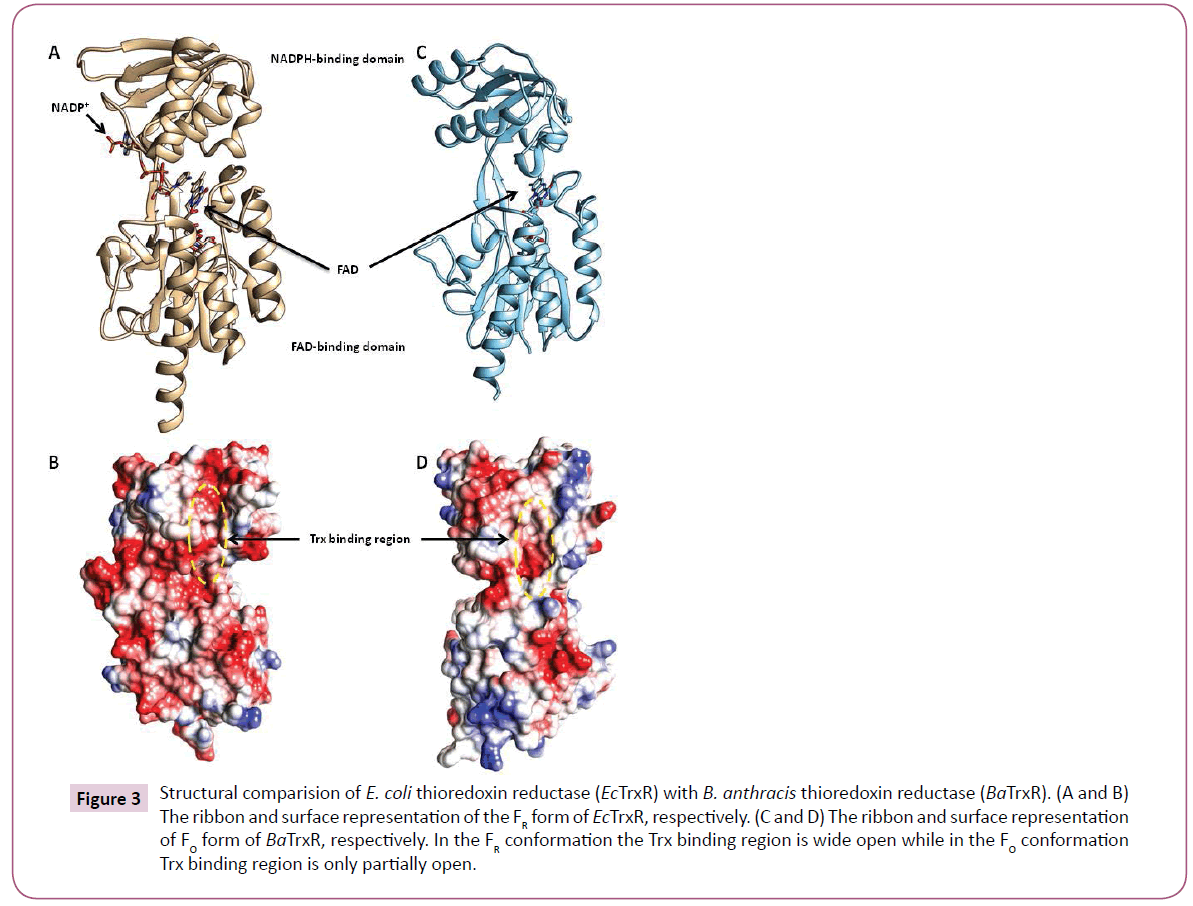
The bacterial thioredoxin reductase is made up of two domains: the NADPH-binding domain and FAD-binding domain (Figure 3). There is ample evidence that during the enzyme catalysis the oxidised conformation (FO) converts to the reduced conformation (FR) by the rotation of the NADPH domain about its axis by 67 degrees. For more details on the conformational changes during binding of Trx in bacterial thioredoxin reductase readers could refer [24]. Most of the structures of TrxR in PDB disclose the FO conformation including the PDB structure of BaTrxR (4FK1) and MtTrxR (28A7). 1F6M is the only X-ray structure from E. coli that shows reduced conformation therefore, the reduced conformation of BaTrxR was modelled using the coordinates of 1F6M.
Figure 3: Structural comparision of E. coli thioredoxin reductase (EcTrxR) with B. anthracis thioredoxin reductase (BaTrxR). (A and B) The ribbon and surface representation of the FR form of EcTrxR, respectively. (C and D) The ribbon and surface representation of FO form of BaTrxR, respectively. In the FR conformation the Trx binding region is wide open while in the FO conformation Trx binding region is only partially open
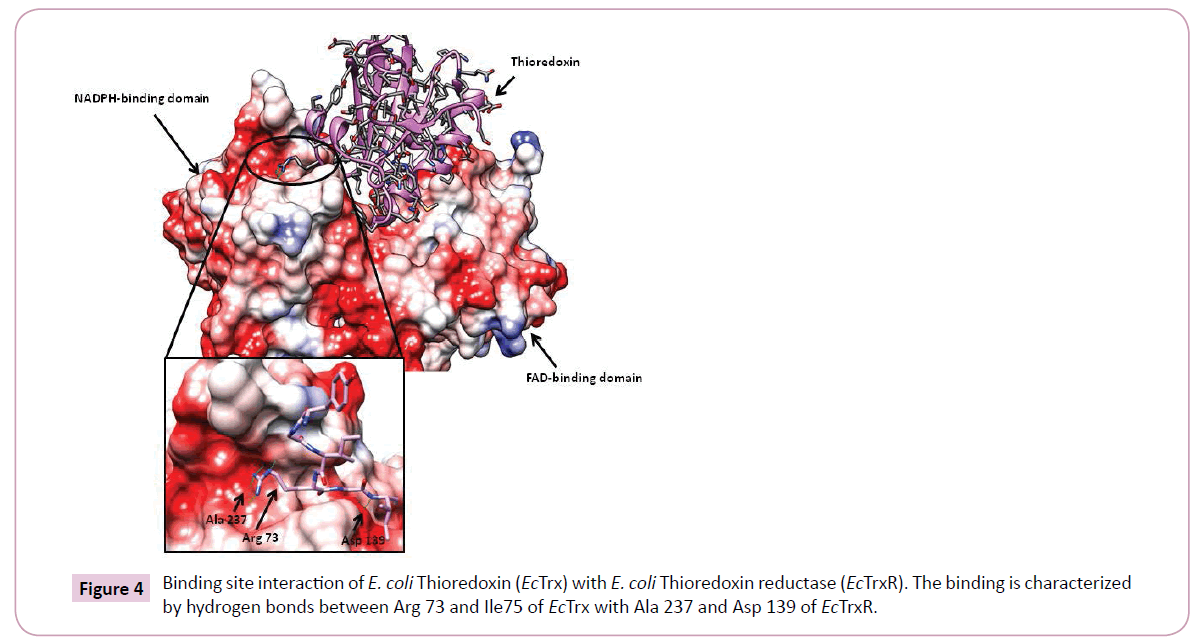
Additionally, the most prominent interactions reported during binding of EcTrx to EcTrxR are observed between the EcTrx loop (Try 70 - Ile75) and complementary pocket on EcTrxR surface. Arg73 side chain of the EcTrx penetrates deep inside the pocket and forms hydrogen bonds with the backbone carbonyl of Ala237 and Arg 130 of the receptor (Figure 4). Other important interactions are seen between Asp 139 side chain of EcTrxR and the backbone amide of EcTrx Ile75. The importance of Arg73 has been demonstrated in a previous study where the loss of activity was observed when Arg was mutated to Gly and Asp [25]. Due to high structural similarity between EcTrxR and BaTrxR a similar binding mode can be expected of BaTrx to BaTrxR characterized by hydrogen bonding to Ala 223 and Asp 137. Therefore, the backbone carbonyl group of Ala 223 and the carboxylate of Asp 137 were selected as the primary target points for potential inhibitors.
Preparation of the protein
The comparison of FO and FR conformations of BaTrxR and EcTrxR shows that FAD domain simply rotates away while binding Trx, without affecting Trx binding. This allowed creating the FR model of BaTrxR by simply removing the FAD domain. Subsequently, Dock Prep tool in UCSF Chimera [26] was used to complete the receptor preparation.
Preparation of the ligands
Compounds obtained after searching the ZINC database were prepared using Structure PrOtonation REcognition System (SPORES) [27]. For this manually added hydrogen atoms in the ligands were first removed and then automatically added by the software. SPORES generates all possible protonation states using rule a rule based combinatorial method. Next, all combinatorial possible tautomers of the input molecules were generated and stored in a multi-mol2 file. Finally the compounds were energy minimized using MMFF94 force field as implemented in openbabel version 2.3.2 [28].
Docking procedure
Docking was carried out using three different docking programs viz. UCSF DOCK version 6.7 (DOCK) [29], Autodock Vina (VINA) [30] and Potein-Ligand ANT System (PLANTS) version 1.2 [31]. The compounds which were found to be common in all three docking results were considered as final predication.
DOCK setup
After the initial preparation of the receptor, DOCK specific steps required for preparing the binding site prior to docking were implemented. The first step in the preparation of the binding site is calculation of the solvent accessible surface area of the receptor, without hydrogen atoms, using the probe radius of 1.4 Å. The negative image of the surface is then created by overlapping spheres using SPHGEN program. The spheres were converted to PDB files for manual selection. The final cluster used for docking had 91 spheres. Subsequently, the grids were calculated in a two step procedure. Firstly, the box around the binding site was constructed with the SHOWBOX program. Secondly, the grids were calculated with the accessory program called GRID using 0.4 grid spacing, 9999 A distance cutoff and a 4r distance dependant dielectric constant. Finally, virtual screening was carried by loading the necessary parameter in the input file with “flexible_ ligand” and “minimize_ligand” parameters turned on.
PLANTS setup
The docking algorithm in PLANTS is based on a class of stochastic optimization algorithms called ant colony optimization. The docking in PLANTS was conducted using the standard settings and PLANTSchemplp scoring function. The binding site was calculated by running the PLANTS in bind mode. Finally, all other necessary parameters were defined in the configuration file and PLANTS was run in screen mode.
VINA setup
For VINA the receptor file and ligands were converted to PDBQT formats using prepare_receptor4.py and prepare_ligand4.py AutoDock scripts. The binding site coordinates were accepted as calculated by PLANTS and the configuration file was prepared with exhaustiveness 8 and cpu value 4.
Results and discussion
Identification of proteins responsible for pathogenicity
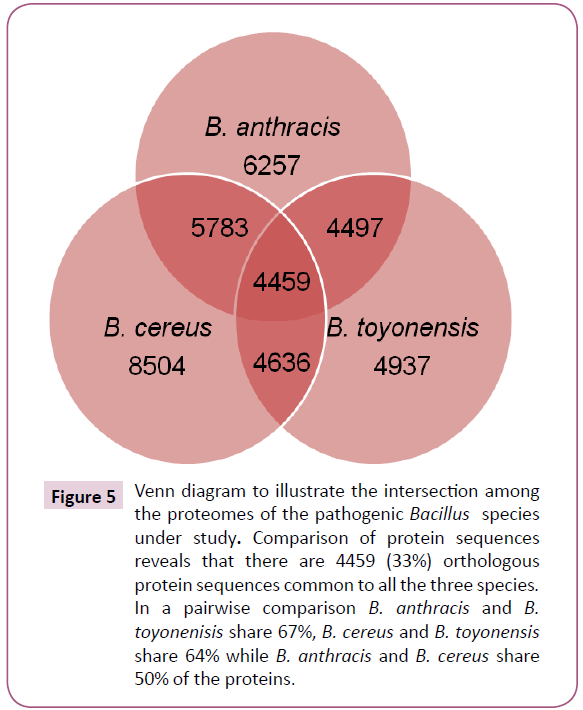
The BLASTP analysis identified 4459 orthologous proteins from 8504, 6257 and 4937 unique protein sequences in B. cereus, B. anthracis and B. toyonensis, respectively (Figure 5). Additionally, in a pairwise comparison, it was observed that 5783 protein sequences were common in B. anthracis and B. cereus (sequences present in pathogenic strains); 4636 were common in B. cereus and B. toyonensis while 4497 in B. anthracis and B. toyonensis. The subtraction of the total number of sequences present in the non-pathogenic strain, B. toyonensis (4937) from the number of sequences present in pathogenic strains (5783) resulted in 846 protein sequences which may play an important role in pathogenicity. The next step in our approach was to filter out sequences similar to human proteins. We found 43 protein sequences to be close enough to human proteins and hence were discarded resulting in 803 proteins from pathogenic strains with no significant sequence similarity to human proteins.
Figure 5: Venn diagram to illustrate the intersection among the proteomes of the pathogenic Bacillus species under study. Comparison of protein sequences reveals that there are 4459 (33%) orthologous protein sequences common to all the three species. In a pairwise comparison B. anthracis and B. toyonenisis share 67%, B. cereus and B. toyonensis share 64% while B. anthracis and B. cereus share 50% of the proteins.
Identification of essential proteins
The proteins involved in virulence and pathogenicity are very important for the existence of pathogens. From 803 non homologous proteins we identified 10 essential proteins from DEG database (Table 1). This information was further exploited to predict novel drug targets.
| UniProt ID | Name of the query protein | Name of the DEG homolog | Homologs in DEG | e-value | Score |
|---|---|---|---|---|---|
| I0D5Y6 | DNA replication protein | DNA biosynthesis protein | 6 | 5.1e-12 | 160 |
| I0D343 | Thioredoxinreductase | Thioredoxinreductase | 5 | 7.5e-14 | 177 |
| I0DAS0 | Ribosomal protein L31 type B | Ribosomal protein L31 | 4 | 1.9e-33 | 199 |
| I0D429 | Membrane protein | Probable conserved integral membrane protein | 3 | 8.2e-12 | 159 |
| I0D0H6 | Carbon starvation protein | Carbon starvation protein | 2 | 2.9e-16 | 200 |
| I0D2E2 | 5-dehydro-2-deoxygluconokinase | Ribokinase-like domain-containing protein | 2 | 2.2e-14 | 182 |
| I0D5Y2 | C-5 cytosine specific DNA methylase | DNA-methyltransferase (cytosine-specific) | 2 | 1.6e-14 | 182 |
| I0CXM4 | Beta-fructofuranosidase | Sucrose-6-phosphate hydrolase/ Beta-fructofuranosidase | 1 | 2.6e-73 | 692 |
| I0D2C8 | Aminoacyl-histidinedipeptidase | Aminoacyl-histidinedipeptidase | 1 | 4.3e-84 | 785 |
| I0D2B8 | Transcriptional regulator, MarR family | MarR family transcriptional regulator | 1 | 3.4e-20 | 228 |
Table 1: Essential proteins of B. anthracis with their corresponding Database of Essential Genes (DEG) homolog, e-value and score. The proteins are arranged in the decreasing order of number of homologs in the DEG database.
Drug target identification and druggability assessment
Druggability is an important feature that describes the properties of proteins which enable interaction with high affinity and specificity to small drug-like molecules. A large number of proteins have structural and physicochemical properties that hinder binding to drugs thus rendering them “undruggable”. Current approaches for identification and evaluation of novel druggable targets are described elsewhere [32]. One of the approaches for evaluating the druggability of a protein is to analyze the sequence homology to known drugs targets. Therefore, 10 essential proteins identified above were aligned to the drug targets in the DB database. Only four proteins were found similar to the binding partners of FDA approved drugs, experimental small molecules or nutraceutical compounds present in DB database. The percentage similarity of the four essential proteins with DB targets, organism name and availability of 3D structural information is shown in Table 2. However, it is important to note that this approach of target identification depends only on the proteins with annotated functions. As such, additional families of druggable proteins might still be identified. Among the four essential proteins thioredoxin reductase was shortlisted as the target of choice for docking analysis due to several reasons mentioned below:
| S.no | UniProt ID | Protein name | DB target | DB organism | % similarity to DB target | Related PDB id | % similarity to PDB |
|---|---|---|---|---|---|---|---|
| 1. | I0CXM4 | Beta-fructofuranosidase | Beta-fructosidase | Thermotogamaritima | 35 (2E-67) | 1UYP | 35 (2E-65) |
| 2. | I0D5Y2 | C-5 cytosine-specific DNA methylase family protein | Modification methylaseHhaI | Haemophilusparahaemolyticus | 27 (1E-16) | 3G7U | 31 (3E-13) |
| 3. | I0D2E2 | 2-keto-3-deoxy-gluconate kinase | 2-keto-3-deoxy-gluconate kinase | Thermusthermophilus | 26 (1E-29) | 2QCV | 60 (6E-141) |
| 4. | I0D343 | Thioredoxinreductase | Thioredoxinreductase | Staphylococcus aureus | 21 (3E-11) | 4FK1 | 100 (0.0) |
Table 2: Non homologous essential proteins of Bacillus anthracis similar to binding partners of FDA approved drugs or experimental compounds.
The thioredoxin system including thioredoxin reductase, thioredoxin and NADPH are critical for DNA synthesis and defence against oxidative stress. Deoxyribonucleotides that are building blocks of DNA and are synthesized by an enzyme called ribonucleotide reductase (RNR), which requires electrons from either thioredoxin or glutaredoxin (Grx) system [glutathione reductase (GR), glutathione (GSH), and Grx]. Interestingly, the Grx system is missing in many pathogenic bacteria such as M. tuberculosis, H. pylori, S. aureus, B. anthracis and B. cereus. This makes Grx-negative pathogens highly susceptible to drugs targeting bacterial TxrR system. Moreover, in other bacteria such as M. tuberculosis [33], S. aureus [34] and N. gonorrhoeae [35], TrxR has been shown to be essential for survival. Furthermore, in contrast to mammalian TrxR, the substrate Trx has to interact directly with the dithiol-disulphide redox centre in the low molecular weight bacterial TrxR, which in inaccessible to the substrate in mammalian TrxR. Therefore, the protein-protein interaction site in B. anthracis TrxR differs from the mammalian counterpart and can be considered for selective targeting. Finally, the availability of the B. anthracis TrxR crystal structure in PDB also impacted our decision to shortlist it for further analysis.
Docking of ligands with Thioredoxin reductase
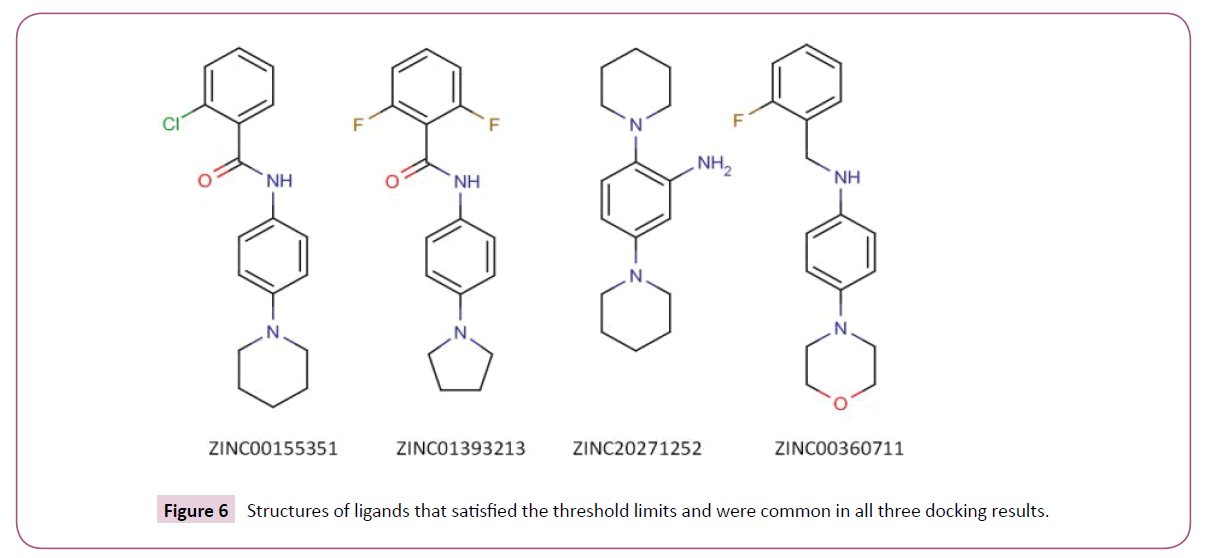
We used three different open source docking programs to predict compounds with high binding affinity to BaTrxR. The importance of Ala 223 and Asp 137 in binding thioredoxin to thioredoxin reductase has been discussed earlier. The formation of hydrogen bonds with the atoms of these two residues by an appropriately placed ligand could lead to ligand occupying the binding cleft thereby blocking the attachment of the native substrate (Trx) to enzyme BaTrxR. Therefore, the compounds that did not form hydrogen bonds with either Ala 223 or Asp 137 residues in the PPI site of the receptor were not considered further for analysis. We determined the hydrogen bond interaction pattern of the docked ligands from FindHbond tool in UCSF Chimera. There were 37 compounds in DOCK, 57 in VINA and 215 in PLANTS that satisfied the hydrogen bond criterion. Before analyzing the common ligands in the three datasets, first we set a threshold docking score as a filter to remove compounds predicted to have low binding affinity. For calculating the threshold score, 10 starting compounds were docked to BaTrxR receptor in all the docking software individually. The threshold score was then set within 10% limit of the average dock score of the starting compounds. Out of the 37 compounds only 20 compounds were above the threshold score of -36 in DOCK dataset. In case of VINA and PLANTS datasets we found that 28 and 65 compounds above the threshold score of -5.1 kcal/mol and -63 respectively. Finally, we found four compounds common in the three docking results. The structure of all the four compounds in 2D format is displayedin (Figure 6) and their Chemical formula, IUPAC names and corresponding PubChem identifiers are in Table 3. The docking scores for each ligand from three different docking programs are shown in (Table 4) while the respective docking poses are displayed in Figure 7. We note that all the four compounds share a similar scaffold which show a hydrogen bond interaction to either Ala 223 or Asp 137 and are a reasonable fit to the pocket in between. The ligands were also stabilized inside the active site through favourable van der Waals and hydrophobic interactions with amino acid residues. Further optimization and testing of the scaffold and its derivatives may provide a more potent and selective BaTrxR inhibitors with better druggability.
| S.No. | ZINC ID | PubChem CID | Chemical Formula | IUPAC Name |
|---|---|---|---|---|
| 1 | ZINC00155351 | 736051 | C18H19ClN2O | 2-chloro-N-(4-piperidin-1-ylphenyl)benzamide |
| 2 | ZINC01393213 | 1480285 | C17H16F2N2O | 2,6-difluoro-N-(4-pyrrolidin-1-ylphenyl)benzamide |
| 3 | ZINC20271252 | 28809802 | C16H25N3 | 2,5-di(piperidin-1-yl)aniline |
| 4 | ZINC00360711 | 834706 | C17H19FN2O | N-[(2-fluorophenyl)methyl]-4-morpholin-4-ylaniline |
Table 3: PubChem Ids, Chemical formulas and IUPAC names of the selected four compounds found common in all three docking programs.
| S.No | ZINC ID | DOCK Scores | PLANTS Scores | VINA binding energies (kcal/mol) |
|---|---|---|---|---|
| 1 | ZINC00155351 | -40.91 | -63.67 | -6.2 |
| 2 | ZINC01393213 | -39.04 | -63.54 | -6.0 |
| 3 | ZINC20271252 | -37.73 | -65.14 | -5.4 |
| 4 | ZINC00360711 | -41.12 | -71.82 | -5.9 |
Table 4: Docking results of the compounds satisfying the threshold scores in the three docking programs.
Conclusions
To conclude, in the present study, proteomes of various pathogenic and non pathogenic strains of B. anthracis and B. cereus were comparatively analysed which resulted in stepwise selection of ten proteins that can be targeted for effective drug design and development. Thioredoxin reductase was finally selected as the most promising target candidate due to essentiality, druggability and availability of X-ray crystallographic structure in PDB. The molecular docking studies with ZINC compounds into the protein-protein interaction site of thioredoxin reductase lead to the identification of novel potential inhibitors sharing a similar scaffold. Future work will focus on further optimization of the scaffold and synthesis of derivatives. We would also like to validate the results of computational analysis in vitro. This study may serve as a template for designing novel inhibitors for pathogens where thioredoxin reductase plays a key in redox homeostasis.
Acknowledgments
We thank Professor Alok Dhawan, Director, Institute of Life Sciences, Ahmedabad University for carefully reading the manuscript.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Chavali AK, D’Auria KM, Hewlett EL, Pearson RD, Papin JA (2012) A metabolic network approach for the identification and prioritization of antimicrobial drug targets. Trends Microbiol 20 (3):113-123.
- Barh D, Jain N, Tiwari S, Parida BP, D’Afonseca V, et al. (2011) A Novel Comparative Genomics Analysis for Common Drug and Vaccine Targets in Corynebacteriumpseudotuberculosis and other CMN Group of Human Pathogens. ChemBiolDrug Des 78 (1):73-84.
- Kaur N, Khokhar M, Jain V, Bharatam PV, Sandhir R, et al. (2013) Identification of Druggable Targets for Acinetobacterbaumannii Via Subtractive Genomics and Plausible Inhibitors for MurA and MurB. ApplBiochemBiotechnol 171 (2):417-436.
- Ahn YY, Lee DS, Burd H, Blank W, Kapatral V (2014) Metabolic network analysis-based identification of antimicrobial drug targets in category A bioterrorism agents. PLoS One 9:e85195.
- Sharma A, Arya DK, Sagar V, Bergmann R, Chhatwal GS, et al. (2013) Identification of potential universal vaccine candidates against group A Streptococcus by using high throughput in silico and proteomics approach. J Proteome Res 12 :336-346.
- Caffrey CR, Rohwer A, Oellien F, Marhofer RJ, Braschi S, et al. (2009) A comparative chemogenomics strategy to predict potential drug targets in the metazoan pathogen, Schistosomamansoni. PLoS One 4: e4413.
- Ariel N, Zvi A, Grosfeld H, Gat O, Inbar Y, et al. (2002) Search for potential vaccine candidate open reading frames in the Bacillus anthracis virulence plasmid pXO1: in silico and in vitro screening. Infect Immun 70: 6817-6827.
- Crowther GJ, Shanmugam D, Carmona SJ, Doyle MA, Hertz-Fowler C, et al. (2010) Identification of attractive drug targets in neglected-disease pathogens using an in silico approach.PLoSNegl Trop Dis 4:e804.
- Mock M, Fouet A (2001) Anthrax.Annu Rev Microbiol 55: 647-671.
- Grunow R, Verbeek L, Jacob D, Holzmann T, Birkenfeld G, et al. (2012) Injection anthrax--a new outbreak in heroin users. DtschArzteblInt 109:843-848.
- Miller JM, Hair JG, Hebert M, Hebert L, Roberts FJ, Jr.,et al.(1997) Fulminating bacteremia and pneumonia due to Bacillus cereus. J ClinMicrobiol35: 504-507.
- Miyata J, Tasaka S, Miyazaki M, Yoshida S, Naoki K, et al.(2013) Bacillus cereus necrotizing pneumonia in a patient with nephrotic syndrome. Intern Med 52: 101-104
- Kotiranta A, Lounatmaa K, Haapasalo M (2000) Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect 2 (2):189-198.
- Hosein IK, Hoffman PN, Ellam S, Asseez TM, Fakokunde A, et al. (2013) Summertime Bacillus cereus colonization of hospital newborns traced to contaminated, laundered linen. JHosp Infect 85:149-154.
- Kumari S, Sarkar PK (2014) In vitro model study for biofilm formation by Bacillus cereus in dairy chilling tanks and optimization of clean-in-place (CIP) regimes using response surface methodology. Food Control 36 (1):153-158.
- Novosad BD, Astley RA, Callegan MC (2011) Role of Toll-like receptor (TLR) 2 in experimental Bacillus cereus endophthalmitis. PLoS One 6 (12):e28619.
- Jimenez G, Blanch AR, Tamames J, Rossello-Mora R (2013) Complete Genome Sequence of Bacillus toyonensis BCT-7112T, the Active Ingredient of the Feed Additive Preparation Toyocerin. Genome Announc 1 (6).
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) BASIC LOCAL ALIGNMENT SEARCH TOOL. Journal of Molecular Biology 215 (3):403-410.
- Zhang R, Ou HY, Zhang CT (2004) DEG: a database of essential genes. Nucleic Acids Res 32:D271-272.
- Ghosh S, Prava J, Samal HB, Suar M, Mahapatra RK (2014) Comparative genomics study for the identification of drug and vaccine targets in Staphylococcus aureus: MurA ligase enzyme as a proposed candidate. J Microbiol Methods 101:1-8.
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, et al.(2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34 (Database issue):D668-D672.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. (2000) The Protein Data Bank. Nucleic Acids Res 28:235-242.
- Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, et al. (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40:D1100-1107.
- Lennon BW, Williams CH Jr, Ludwig ML (2000) Twists in catalysis: alternating conformations of Escherichia coli thioredoxinreductase. Science 289 (5482):1190-1194.
- Negri A, Rodriguez-Larrea D, Marco E, Jimenez-Ruiz A, Sanchez-Ruiz JM, et al. (2010) Protein-protein interactions at an enzyme-substrate interface: characterization of transient reaction intermediates throughout a full catalytic cycle of Escherichia coli thioredoxinreductase. Proteins 78 (1):36-51.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. (2004) UCSF Chimera--a visualization system for exploratory research and analysis. J ComputChem 25 (13):1605-1612.
- ten Brink T, Exner TE (2009) Influence of protonation, tautomeric, and stereoisomeric states on protein-ligand docking results. J ChemInf Model 49: 1535-1546.
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, et al. (2011) Open Babel: An open chemical toolbox. J Cheminform 3:33.
- Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT, et al. (2015) DOCK 6: Impact of new features and current docking performance. J ComputChem 36 (15):1132-1156.
- Trott O, Olson AJ (2010) AutoDockVina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J ComputChem 31 (2):455-461.
- Korb O, Stützle T, Exner T (2007) An ant colony optimization approach to flexible protein–ligand docking. Swarm Intell 1 (2):115-134.
- Egner U, Krätzschmar J, Kreft B, Pohlenz H-D, Schneider M (2005) The Target Discovery Process. ChemBioChem 6:468-479.
- Sassetti CM, Boyd DH, Rubin EJ (2003) Genes required for mycobacterial growth defined by high density mutagenesis. MolMicrobiol 48:77-84.
- Uziel O, Borovok I, Schreiber R, Cohen G, Aharonowitz Y (2004) Transcriptional regulation of the Staphylococcus aureusthioredoxin and thioredoxinreductase genes in response to oxygen and disulfide stress. J Bacteriol 186:326-334.
- Potter AJ, Kidd SP, Edwards JL, Falsetta ML, Apicella MA, et al. (2009) Thioredoxinreductase is essential for protection of Neisseria gonorrhoeae against killing by nitric oxide and for bacterial growth during interaction with cervical epithelial cells. J Infect Dis 199 (2):227-235.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences