Structure Homology Modeling of Thaumetopoein, an Urticating Protein from Thaumetopoea pityocampa Schiff, Using SWISS-MODEL Workspace
Djillali Tahri,Mohammed Seba and Khedidja Benarous
Djillali Tahri, Mohammed Seba and Khedidja Benarous*
Biology Department, Amar Telidji University, Laghouat, Algeria
- *Corresponding Author:
- Khedidja Benarous
Biology Department, Amar Telidji University
Laghouat, Algeria
E-mail: k.benarous@lagh-univ.dz
Received date: November 05, 2015; Accepted date: November 07, 2015; Published date: November 15, 2015
Citation: Tahri D, Seba M, Benarous K. Structure Homology Modeling of Thaumetopoein, an Urticating Protein from Thaumetopoea pityocampa Schiff, Using SWISS-MODEL Workspace. Chem Inform. 2015, 1:2.
Abstract
Thaumetopoea pityocampa Schiff. or pine caterpillar, is a phyto; xylophagous lepidopteran. For protection against predators, this caterpillar produce an urticating hairs (setae) inducing increasingly cutaneous reactions in animals and humans (erusim). Caterpillar hairs contain a specific protein which has an urticating properties named thaumetopoein. In this study, we started with phylogenetic reconstruction of some chemosensory proteins family with thaumetopoein sequence, then we proceded to homology modeling to elucidate the threedimensional structure of this allergic protein and we got a more reliable model based on the results obtained (QMEAN score=0.73, Z-score=-0.75 with 0.940 a high global relative model SELECTpro confidence score) which is validated with two other models resulting from other web servers such as Phyre2 and MetaMQAPII.
Keywords
Thaumetopoea pityocampa Schiff; Thaumetopoein; Homology Modeling; Pine Caterpillar
Background
Thaumetopoea pityocampa Schiff., or pine caterpillar, is a “phenomenal” insect. The term comes from the Greek cámpa (caterpillar), pítys (pine), poieo (does), tháuma (wonders) [1]. When developing larvae, is taking place on the back of the caterpillar pine processionary, a producer apparatus urticating hairs, protectors caterpillars [2]. These hairs of the pine processionary caterpillar (Thaumetopoea pityocampa Schiff.) causes dermatological reactions in humans by contact with its irritating larvae hairs known as erucism, the pathogenic effects are not limited to the skin but extend to the eyes and, more rarely, to the respiratory system [1,3]. A various techniques demonstrated the existence of a specific protein fraction in caterpillar hairs which has urticating properties and which we have called thaumetopoein [3-5]. The structure and properties of this protein have not been fully elucidated, knowing that Rodriguez-Mahillo et al. have published in 2012 the protein sequence in under NCBI ID: CCJ09295 and EMBL EBI ID: HE962022. In this study, we are trying using bioinformatics and modeling tools to give a prediction model of thaumetopoein structure and its properties.
Materials and Methods
Database research
The amino acid sequence of this protein was then obtained in NCBI (The National Center for Biotechnology Information), using protein-BLAST research web tool (NCBI ID: CCJ09295, EML EBI ID: HE962022); a comparison of this sequence with those of RCSB PDB (Protein Data Bank) was performed. Sequences of proteins that are more like the query sequence, were selected for phylogenetic reconstruction (Table 1).
| 2GVS | Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria |
| 1KX8 | Structure and ligand binding study of a moth chemosensory protein |
| 2JNT | Structure of Bombyx morichemosensory protein 1 in solution. |
| 2OAP | Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism |
| 5EAT | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase |
| 5EAS | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. |
| 1X6C | Solution structures of the SH2 domain of human protein-tyrosine phosphatase SHP-1 |
| 2ZZF | The structure of alanyl-tRNA synthetase with editing domain. |
Table 1: Sequences taken from the results obtained by protein BLAST.
Alignment and phylogenetic reconstruction
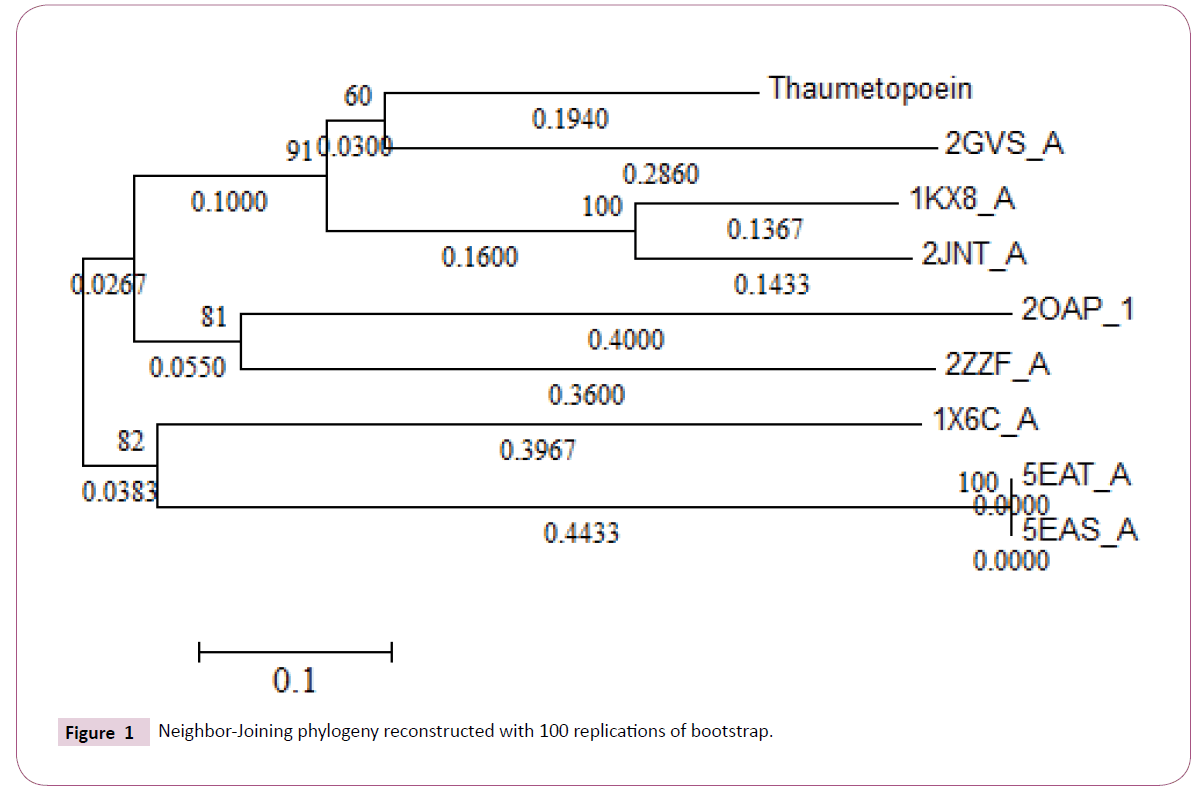
Selected sequences were aligned with COBALT (Constraint-based Multiple Protein Alignment Tool) (https://www.ncbi.nlm.nih.gov/ tools/cobalt/). With the result of alignment we preceded to phylogenetic reconstruction by MEGA software using p-distance calculations for branches length and Neighbor-Joining statistical method with 100 replication of bootstrap.
Protein homology modeling
With modeling tool by Swiss Model (https://swissmodel.expasy.org/workspace/) [6-8] and the alignment results and theSwiss Model , it was be able to make two models of this protein, each one was made following an alignment with 2GVS and 1KX8 respectively. A comparison of the two models thus obtained was made using FATCAT (Flexible structure AlignmenT by Chaining Aligned fragment pairs allowing Twists) pairwise alignment (https://fatcat.burnham.org/) to verify the degree of similarity between these two models. For model quality estimation and validation, in addition to QMEAN (Qualitative Model Energy ANalysis) which is a default option in Swiss Model for the estimation of the best reliable model quality, we used SELECTpro web tool (https://www.igb.uci.edu/~baldig/ selectpro.html) who takes the amino acid sequence of a protein and the backbone coordinates of a corresponding model as input and calculates a total pseudo-energy. This value is normalized to the 0.0 to 1.0 confidence scale. Higher scores indicate higher relative model confidence. We also conducted to predict the structure with other web servers: Phyre2 (https://www.sbg. bio.ic.ac.uk/phyre2/html/page.cgi?id=index) and MetaMQAPII (https://genesilico.pl/toolkit/unimod?method=MetaMQAPII) as validation methods.
Results and Discussion
Database research
After comparing the query sequence with protein BLAST, sequences were selected (Table 1) according to the parameters; max score, total score, E-value and percent identity.
Alignment and phylogenetic reconstruction
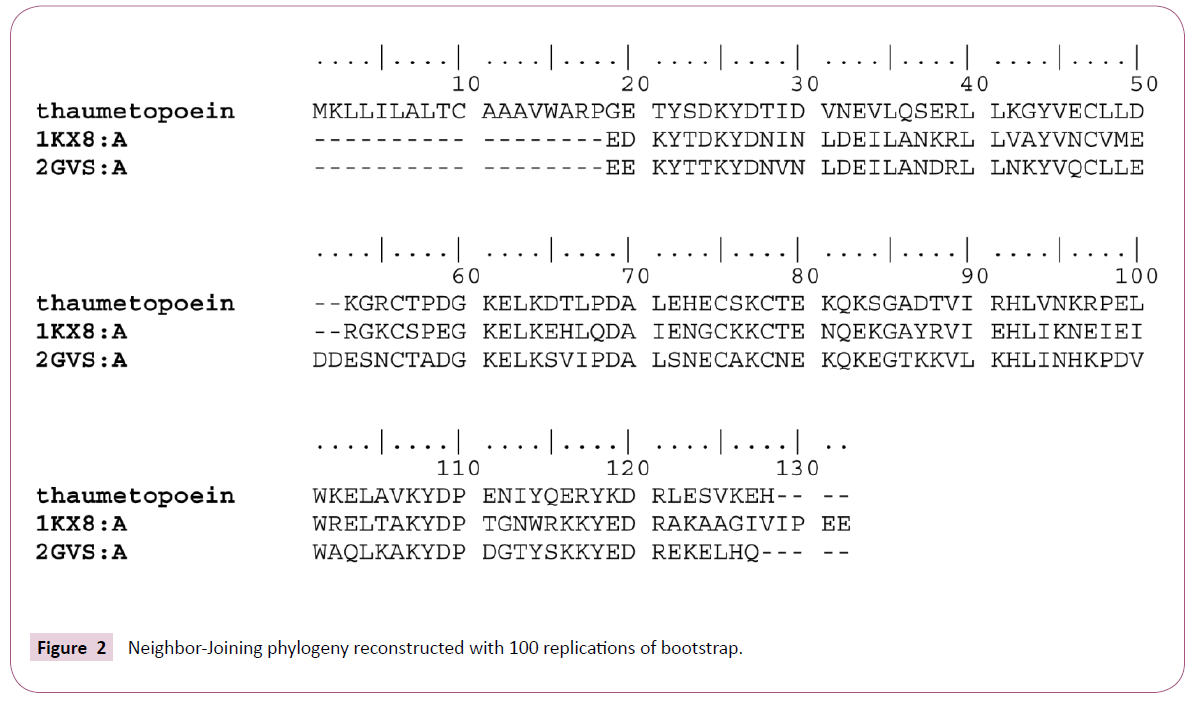
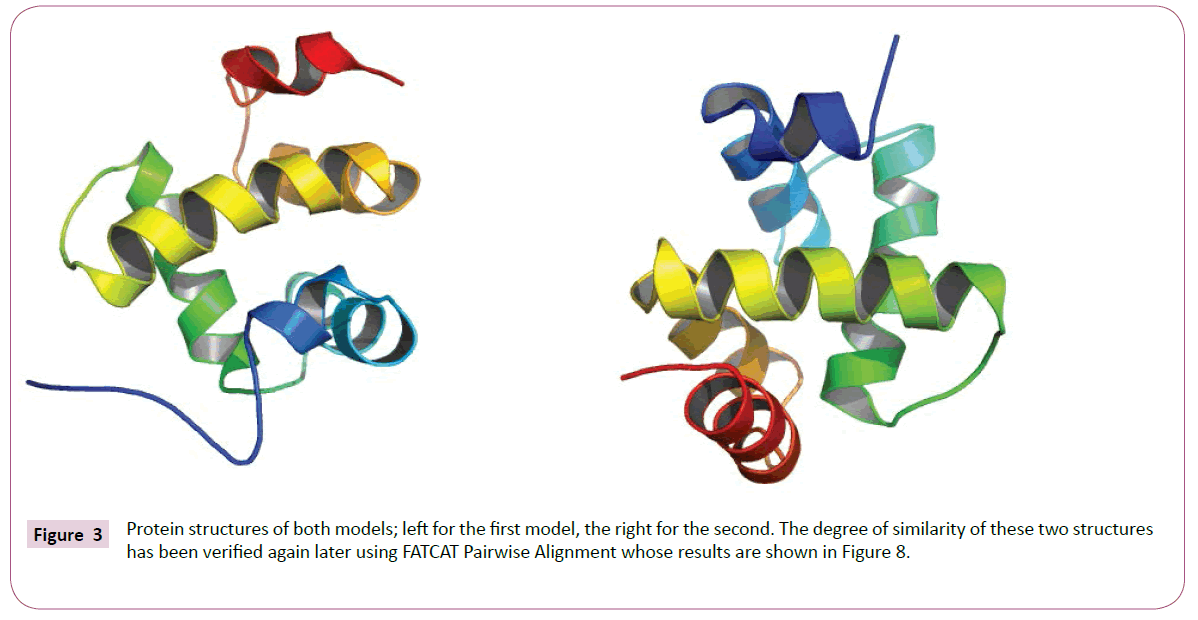
Multiple alignments of the sequences by COBALT was followed by a calculation of p-distances with MEGA software then reconstructing phylogenetic tree according to the neighborjoining method (Figure 1). From the phylogenetic tree we can consider the presence of four monophyletic groups which show an inter-resemblance except in the case of 2GVS that was regarded as an in group, because thaumetopoein, 1KX8 and 2JNT are chemosensory proteins and later we find that the two models that were made from the homology modeling with 1KX8 and 2GVS (Figure 2) are reliable structure with no greater difference (Figures 3-6).
Protein homology modeling
Both models were produced according to the initial alignment either with 2GVS or 1KX8 are based on links shown by the phylogenetic tree, whose protein structures are shown in Figure 3. The characteristics of the two models are shown in Figure 4. In the Figure 5 we have the graphical representation for predicted local error of structures of the two models.
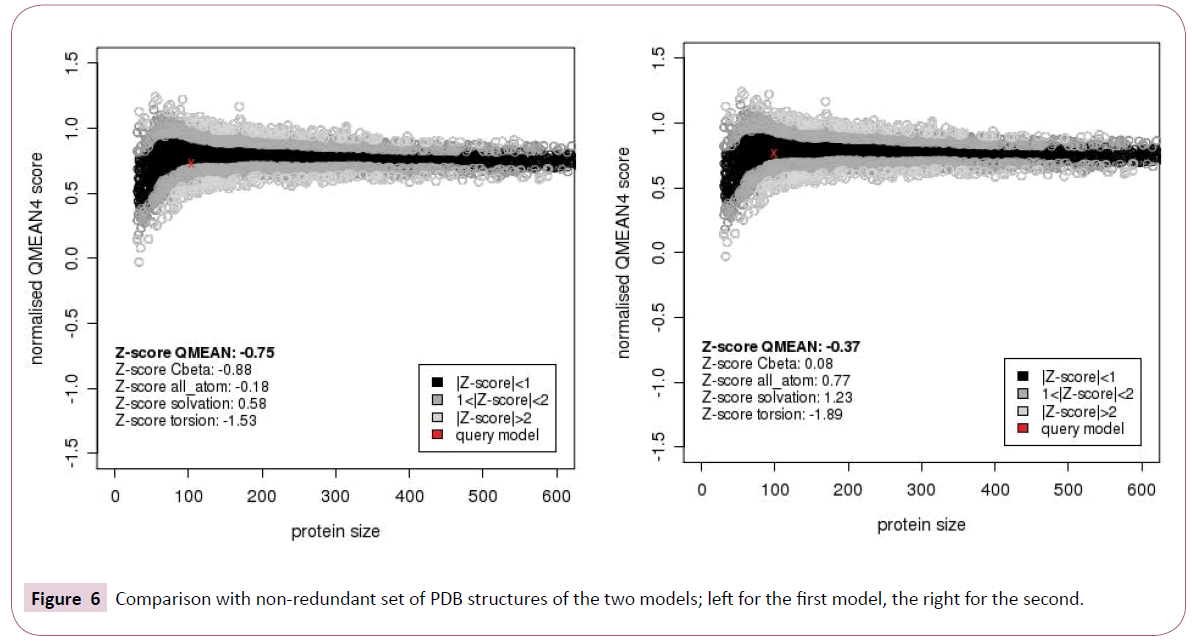
Both models obtained after homology modeling of the query sequence of thaumetopoein and both 2GVS and 1KX8 are sized 104 and 99 amino acids respectively. While taking into account the value of QMEAN (Qualitative Model Energy ANalysis) Z-score (Figure 4) that is no large difference compared to the Z-scores of which the values that are not too similar (-0.75 and -0.37 respectively). In addition, the second model shows less predicted local errors (Figure 5) and more estimated absolute model quality (Figure 6). The results of estimated absolute model quality by comparison with non-redundant set of PDB structures of the two models are shown in the Figure 6.
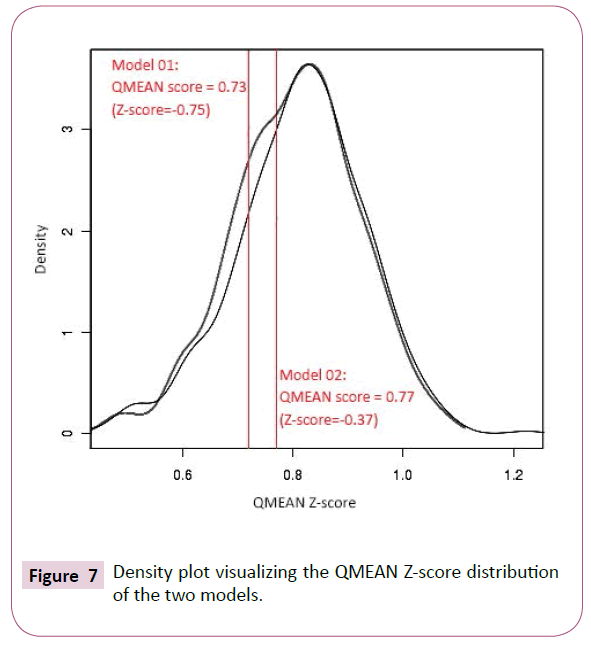
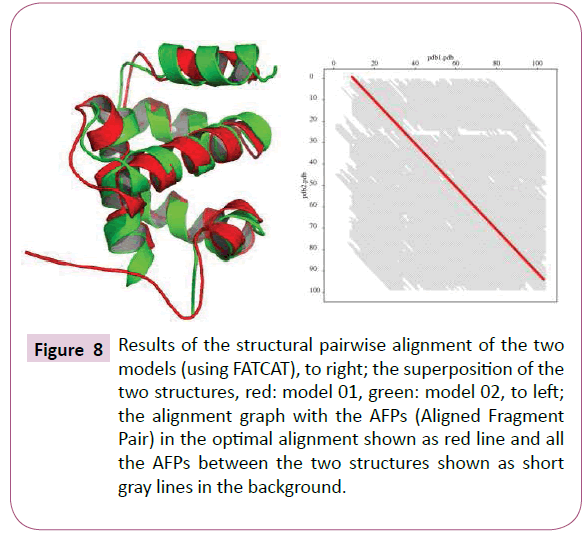
From density plot (Figure 7), we see a slight difference and more significant structural reliability of the first model (which is validated using SELECT pro web tool with 0.940 a high global relative model confidence score) [9]. According to Benkert et al. the "good" models have a QMEAN Z-score in between -0.65 and the "medium" quality models in between -1.75 [10]. This small difference was demonstrated by pairwise alignment FATCAT web tool, resulting a significant similarity with P-value of 1.04e-10, given that the structure alignment has 94 equivalent positions with an RMSD of 2.63 (Figure 8) [10,11]. The other two models so obtained by Phyre2 (85% of residues modelled at more than 90% confidence, (Figure 9) [12] and MetaMQAPII (with an interesting GDT TS=61.058 and RMSD=2.851) [13] showed great significant similarity with the first two SWISSMODEL models.
Figure 8: Results of the structural pairwise alignment of the two models (using FATCAT), to right; the superposition of the two structures, red: model 01, green: model 02, to left; the alignment graph with the AFPs (Aligned Fragment Pair) in the optimal alignment shown as red line and all the AFPs between the two structures shown as short gray lines in the background.
Conclusion
Like all proteins Insect pheromone-binding family A10/OS-D (PFAM ID: PF03392), thaumetopoein appears as a small hydrophobic helical protein. However, resulting models of homology modeling using three web servers (SwissModel, Phyre2, MetaMQAPII) show a high structural reliability and significant similarity, which leads us to think about the study of physico-chemical properties in relation with the roles and biological effects, considering the rapid propagation of the pine processionary moth and its extensive invasion every year.
References
- Bonamonte D, Foti C, Vestita M, Angelini G (2013) Skin Reactions to pine processionary caterpillar Thaumetopoea pityocampa Schiff. ScientificWorldJournal 2013: 867431.
- Novak F, Lamy M (1987) Etude ultrastructurale de la glande urticante de la chenille processionnaire du pin, thaumetopoea pityocampa schiff. (lepidoptere: thaumetopoeidae). Int J Insect MorphoL & Ernbryol 16:263-270.
- Lamy M, Pastureaud MH, Novak F(1986) Thaumetopoein: an urticating protein from the hairs and integument of the pine processionary caterpillar (thaumetopoea pityocampaschiff., lepidoptera, thaumetopoeidae). Taxiton 24:347-356.
- Novak F, Pelissou V, Lamy M (1987) Comparative morphological, anatomical and biochemical studies of the urticating apparatus and urticating hairs of some lepidoptera: Thaumetopuea pityocampa schiff., Th. processionea L. (lepidoptera, thaumetopoeidae) and Hylesia Metabus Cramer (lepidoptera, saturniidae). Camp Biochem Physio 88A: 141-146.
- Rodriguez-Mahillo AI, Gonzalez-Muñoz M, Vega JM, López JA, Yart A, et al. (2012) Setae from the pine processionary moth (Thaumetopoea pityocampa) contain several relevant allergens. Contact Dermatitis 67: 367-374.
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195-201.
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res 31: 3381-3385.
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714-2723.
- Randall A, Baldi P (2008) SELECTpro: effective protein model selection using a structure-based energy function resistant to BLUNDERs. BMC Struct Biol 8: 52.
- Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27: 343-350.
- Ye Y, Godzik A (2003) Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19 Suppl 2: 246-255.
- Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363-371.
- Kalman M, Ben-Tal N (2010) Quality assessment of protein model-structures using evolutionary conservation. Bioinformatics 26: 1299-1307.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences