Structural Features on Polymorphism of Diclofenac Acid
Borges RS,Carvalho ES, Gabriel S Cabral, Taina G Barros, Carlos A L Barros, Agnaldo S Carneiro
Borges RS, Carvalho ES, Gabriel S Cabral, Tainá G Barros, Carlos A L Barros, Agnaldo S Carneiro
Faculdade de Farmácia, Instituto de Ciências da Saúde, Universidade Federal do Pará, Belém, PA, Brazil
- *Corresponding Author:
- Rosivaldo S. Borges
Faculdade de Farmacia, Instituto de Ciencias da Saude
Universidade Federal do Para, 66075-110, Belem, PA
Brazil
Tel: +55 91 3201 7202
Fax: +55 91 3201 7201
E-mail: rosborg@ufpa.br
Received date: June 30, 2015; Accepted date: September 08, 2015; Published date: September 11, 2015
Citation: Borges RS, Carvalho ES, Gabriel S Cabral, et al. Structural Features on Polymorphism of Diclofenac Acid. Chem Inform. 2015, 1:1.
Abstract
Objective:
In this report, three dimensional structure of lanosterol 14-α demethylase (CYP51) protein of Candida albicans was modeled and used for docking studies with eighteen synthetic triazole derivatives.
Methods:
The model was generated by multiple threading alignment and iterative structure assembly simulation by I-TASSER and validated by Ramachandran plot using PROCHECK and MolProbity servers. The eighteen 1,2,3-triazole compounds were previously synthesized using two different synthetic routes by our research group in the search of new antifungal agents. The binding affinity and interaction of all triazole compounds with predicted model of CYP51 were determined by AutoDock Vina.
Results:
I-TASSER generated five models in which Model 1 was selected as best predicted structure on the basis of C-score 1.30, TM-score 0.89 ± 0.07, and RMSD 4.7 ± 3.1 Å. It showed 89.2% residues in most favoured region and 7.1% residues in additional allowed regions. Docking results showed that all the derivatives have good binding energy in the range -9.8 to -7.4 kcal/mol. Moreover, compound NT- 03 (-9.8 kcal/mol) was found as best inhibitor as it binds nicely into the active site of the enzyme.
Conclusion:
The study suggests that I-TASSER is very easy and reliable tool for three dimensional structure predictions. The N-atom in quinoline ring of compound NT- 03 involved in the additional interaction with predicted model of CYP51. Finally, I-TASSER might be used as an important tool in drug designing process in search of more effective therapies.
Keywords
Diclofenac acid; DFT; Hydrogen bond; Structure; Conformation; Polymorphism
Introduction
Diclofenac acid or 2-[(2,6-dichloro-phenyl)-amino]-phenylacetic acid is an excellent nonsteroidal anti-inflammatory drug (NSAID) [1] used in the treatment of pain disorders. It has numerous solid forms, including various diclofenac salts. Among them, an exemplary of diclofenac acid crystal forms including one monoclinic form which is recrystallized from methanol by slow evaporation, and another which is recrystallized from acetone [2]. Diclofenac acid also exists in an orthorhombic form, which is recrystallized from hot methanol by slow evaporation [3]. Several forms of diclofenac salts include potassium diclofenac dihydrate [4], sodium diclofenac tetrahydrate [5], and sodium diclofenac pentahydrate [6]. Thus, the polymorphism of diclofenac depends on the recrystallization conditions [7,8].
In fact, the study of polymorphism in pharmaceutical substances is important because of possible changes in their solubility, dissolution and shelf life [9]. Therefore, it is important to characterize both pharmaceutical and theoretical properties in their solid state and formulations, especially those substances dispersed or embedded in a polymer matrix and different solvent, to establish that they still preserve the required pharmacological properties, including their chemical and physical properties and biological activities.
Actually, the use of computational methods is an indispensable tool for the understanding of the physical and chemical properties of molecules, mainly related to its use in pharmaceutical systems. In this work, a theoretical study of diclofenac acid is performed to understand the intramolecular or intermolecular interactions, providing information about the variation of the most stable polymorphic structures of diclofenac acid.
All calculations were performed with the Gaussian 03 molecular package [10]. Prior to any DFT calculations, the structure of diclofenac acid was submitted to PM3 [11] geometry conformational search. Afterwards, the PM3 geometry was fully optimized with the B3LYP hybrid density functional theory [12,13] using the 6-31G(d,p) basis sets [14,15]. Only the most stable conformation for a given structure was used and all of them are free from negative frequencies. The energy barrier (ΔE) was calculated as the energy difference between the structures of lower energies (2-5) and the respective the structure of higher energy (1) (Eq. 1).
ΔE=Ef -Ei (Eq. 1)
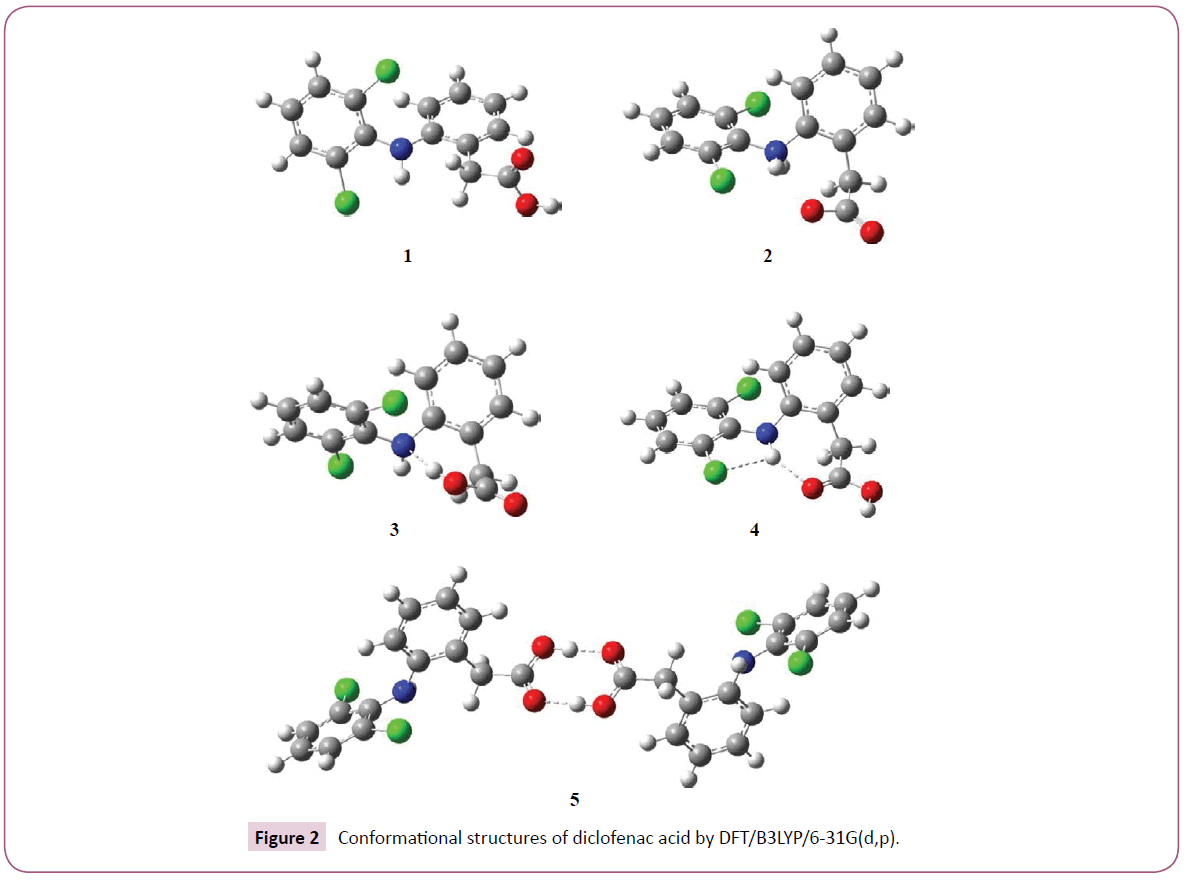
summarizes the ΔE and their corresponding conformational structures obtained by using the hybrid Density Functional Theory B3LYP and 6-31G(d,p) basis set. We have found five main structures named gas phase (1) zwitterion (2), acid-base interaction (3), orthorhombic structure (4), and dimer (5) due to their energy or conformational structures.
In drug development different forms of a pharmaceutical substance may display different physical and chemical properties. Thus, the pharmaceutical substances may change to different polymorphs, solvates or pseudo-polymorphs, de-solvates or amorphous materials after standard pharmaceutical processes such as crystallization, milling, freeze drying, spray drying and solid dispersion, which may change their pharmaceutical activities [9]. In fact, every structure has different energies among their diverse physical forms. Therefore, the pharmaceutical industry requires a strategy to characterize polymorphic drugs and produce drugs of consistent quality and the theoretical methods can be used for a better understanding.
Concerning energy barriers (ΔE) (Figure 1), the B3LYP/6- 31G(d,p) basis set two groups were separated using interactions mainly hydrogen bond: intermolecular and intramolecular. The zwitterion (2) formation, where the ΔE values of 49.68 kcal/mol can be unfavorable. The high values are due to a low basicity on nitrogen biphenyl. In addition, two chloro on ortho position of phenyl ring decrease the nitrogen basicity. In fact, the hydrogen bond interaction on acid-base structure (3) had shown a low ΔE values of -0.17 kcal/mol when compared with gas phase structure (1).
Nevertheless, the increase of hydrogen bonding gives structure more stable. Therefore, the orthorhombic structure (4) and dimer (5) have shown the lower ΔE values of -5.59 and -20.42 kcal/mol, respectively, where the N-H works like an acid group and carbonyl acting as an electron donating group.
However, the orthorhombic structure (4) has more functional groups involved with molecular interaction, forming a more rigid structure than dimer (5). This behavior restricts other possible conformations. Further, dimer (5) could have a difficult solvation mainly due to its high molecular weight. Hence, the high molecular structure may be related with an increase of intermolecular repulsion and because of its different free dihedral groups, other conformations should be possible. Therefore the dimer structure (5) can be more flexible. In Figure 2, all intramolecular and intermolecular interactions can be observed.
Furthermore, there have been reports that the physical and chemical properties of diclofenac depends on the acidity of the conditions to which it is exposed. The solubility of diclofenac is substantially governed by the pH of the surrounding solution [16,17]. It can undergo an intramolecular cyclization under the acidic conditions found in gastric juices [18]. Herein, our theoretical results mainly for the orthorhombic structure (4) are in agreement to its crystallographic structure obtained by using of X-ray powder diffraction.
Conclusion
The theoretical study shows that the most stable conformational structure exhibited the lowest energy barriers. This property can be attributed to increase of intermolecular when compared to intramolecular interactions. The increase of hydrogen bonding gives more stable structures. Orthorhombic structure has more functional groups involved with molecular interaction than dimer structure. These results explain the probable more involvement of oxygen, halogen and nitrogen groups on intramolecular interaction of stable conformation.
References
- Skoutakis VA, Carter CA, Mickle TR, Smith VH, Arkin CR, et al. (1988) Review of diclofenac and evaluation of its place in therapy as a nonsteroidalantiinflammatory agent. Drug IntellClin Pharm 22: 850-859.
- Castellari C, Ottani S (1997) Two monoclinic forms of diclofenac acid. ActaCryst C53: 794-797.
- Jaiboon N, Yosin K, Ruangchaithaweesuk S, Chaichit N, Thutivoranath R, et al. (2001) New orthorhombic form of 2-[(2,6-dichlorophenyl)amino]-benzeneacetic acid (Diclofenac acid). Anal Sci 17: 1465–1466.
- Fini A, Garuti M, Fazio G, Alvarez-Fuentes J, Holgado MA (2001) Diclofenac salts. I. Fractal and thermal analysis of sodium and potassium diclofenac salts. J Pharm Sci 90: 2049-2057.
- Fini A, Fazio G, Alvarez-Fuentes J, Fernàndez-Hervàs MJ, Holgado MA (1999) Dehydration and rehydration of a hydrate diclofenac salt at room temperature. Int J Pharm 181: 11-21.
- Muangsin N, Prajaubsook M, Chaichit N, Siritaedmukul K, Hannongbua S (2002) Crystal structure of a unique sodium distorted linkage in diclofenac sodium pentahydrate. Anal Sci 18: 967-968.
- Fini A, Fazio G, Fernández Hervás MJ, Holgado MA, Rabasco AM (1996) Factors governing the dissolution of diclofenac salts. J Pharm Sci 4: 231-238.
- O'Connor KM, Corrigan OI (2001) Comparison of the physicochemical characteristics of the N-(2-hydroethyl)pyrrolidine, diethylamine and sodium salt forms of diclofenac. Int J Pharm 222: 281–293.
- Hosokawa K, Goto J, Hirayama N (2005) Predicition of solvents suitable for crystallization of small organic molecules. Chemical and Pharmaceutical Bulletin 53: 1296-1299.
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al. (2004) Gaussian 03, Revision C.02 Gaussian, Inc., Wallingford CT.
- Stewart JJ (1989) Optimization of Parameters for Semi-Empirical Methods I-Method. J Comp Chem 10: 209-220.
- Kohn W, Becke AD, Parr RG (1996) Density Functional Theory of Electronic Structure. J PhysChem 10: 12974-12980.
- Becke AD (2014) Perspective: Fifty years of density-functional theory in chemical physics. J ChemPhys 140: 18A301.
- Lee C, Yang WR, Parr GR (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B 37: 785-789.
- Hehre WJ, Radom L, Schleyer PVR, Pople JA (1986) Ab Initio Molecular Orbital Theory. John Wiley and Sons: New York 7:1-379.
- Palomo ME, Ballesteros MP, Frutos P (1999) Analysis of diclofenac sodium and derivatives. J Pharm Biomed Anal 21: 83-94.
- Tudja P, Khan MZ, Mestrovic E, Horvat M, Golja P (2001) Thermal behaviour of diclofenac sodium: decomposition and melting characteristics. Chem Pharm Bull 49: 1245-1250.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences